CPC Definition - Subclass C12N
This place covers:
- Microorganisms (e.g. protozoa, bacteria, fused plant cells, hybridomas, viruses, animal cells or tissue, stem cells, tumour cells) and enzymes or proenzymes and compositions containing microorganisms and enzymes or proenzymes.
- Processes for preparing, activating, inhibiting, separating, or purifying enzymes.
- Treatment of microorganisms or enzymes with electrical or wave energy.
- Processes of reproducing, maintaining, or preserving microorganisms or compositions thereof.
- Processes of preparing or isolating a composition containing microorganisms.
- Preparing mutants and screening processes therefor.
- Processes of fusing two or more cells to each other.
- Recombinant DNA-technology including:
- Processes for manipulating genetic material;
- Processes of preparing, isolating and purifying nucleic acids;
- Methods for the introduction of genetic material into microorganisms using vectors or other expression systems, using microencapsulation, using microinjection, and other ways;
- Methods of regulating gene expression;
- Non-coding nucleic acid sequences, e.g. Promoters, operators, enhancers, suppressors, silencers, locus control regions, antisense nucleic acids, and aptamers, used in regulating gene expression or in other recombinant DNA technology related methods.
- Genes, per se; and vectors and expression systems, per se.
- Media for supporting or sustaining the growth of microorganisms.
In subclasses C12M - C12Q, in the absence of an indication to the contrary, classification is made in the last appropriate subclass of subclasses C12M - C12Q.
Multiple Classification
- Compositions, physical forms, methods of application of specific materials of the use of single compounds or compositions as biocides, pest repellants, pest attractants, pesticides, herbicides or plant growth regulators are further classified in subclass A01N.
- Biocidal, pest repellant, pest attractant, or plant growth regulatory activity of chemical compounds or preparations is further classified in A01P.
- Therapeutic activity of compounds containing microorganisms, single cell proteins, or enzymes, is further classified in subclass A61P.
- Uses of cosmetics or similar toilet preparations containing microorganisms or enzymes are further classified in subclass A61Q.
It is desirable to add subclass C12R for microorganisms which are considered to be of interest for search.
This place does not cover:
Microbiological testing media |
Examples of places where the subject matter of this place is covered when specially adapted, used for a particular purpose, or incorporated in a larger system:
New breeds of multicellular plants, e.g. non-transgenic plants, and processes of obtaining these plants | |
New breeds of multicellular animals, e.g. transgenic animals, and processes of obtaining these animals | |
Compositions and use of the compositions and compounds for preservation of bodies of humans or animals or parts thereof | |
Compositions and use of the compositions and compounds for preservation of plants or parts thereof | |
Biocides, pest repellents or attractants or plant growth regulators containing microorganisms, viruses, microbial fungi, enzymes, fermentates, or substances produced by, or extracted from, microorganisms or animal material | |
Bakery products which may contain microorganisms or enzymes | |
Foods or foodstuffs containing microorganisms or enzymes | |
Body treating or pharmaceutical preparations containing microorganisms or enzymes | |
Medicinal preparations containing nucleic acids | |
Medicinal preparations containing genetic material which is inserted into cells of the living body to treat genetic diseases; Gene therapy | |
Bandages, dressings or absorbent pads for physiological fluids containing microorganisms | |
Bandages, dressings or absorbent pads for physiological fluids containing enzymes | |
Biological compost | |
Organic fertilizers containing added bacterial cultures, mycelia or the like | |
Enzyme containing detergent compositions |
Attention is drawn to the following places, which may be of interest for search:
Preservation of excised living parts of the human or animal body | |
Food compositions | |
Chemical aspects of, or use of materials for, bandages, dressings, absorbent pads or surgical articles | |
Nucleic acids not used in recombinant technology and their chemical preparation | |
Compositions, characterized by the use of bacteria, which are used to enhance recovery of hydrocarbons from underground formations |
In this subclass, with the exception of group C12N 5/06, in the absence of an indication to the contrary, classification is made in the last appropriate place.
In this subclass, viruses, human, animal, or plant cells, protozoa, tissues, and unicellular algae are considered as microorganisms.
Overview of relevant orthogonal Indexing symbols:
Indexing symbols intended for the nutritive components of culture media in combination with C12N 5/0018 (generic media) or C12N 5/06 and subgroups (specific media)—but effects beyond nutrition are not excluded. There is some aspect of classification by pathway in that C12N 2500/10 and subgroups cover metals as well as metal chelators. Note that C12N 2500/25 substitutes for any combination of C12N 2500/05 (for selenium), C12N 2500/24 (transferrin/iron) and C12N 2501/33 (insulin). An example of "undefined extract" is Bovine Pituitary Extract (BPE), C12N 2500/84; serum is not indexed as undefined extract since codes are provided for its explicit absence. C12N 2500/02 codes for explicitly low (or high) O2 pressure, not for the "usual" 5% CO2. Antibiotics are not foreseen in the scheme.
Indexing symbols intended for biologically active agents in culture and differentiation processes in combination with C12N 5/06 and subgroups or C12N 5/0018. Indexation is made at the most relevant place, taking account of the biological pathway involved and not the chemical structure, unlike apparently similar hierarchies such as those found in C07K 14/00 or A61K 38/00; e.g.OKT3 antibody C12N 2501/515, staurosporine C12N 2501/727 (tyrosine kinase inhibitor), KAAD-cyclopamine C12N 2501/41 (interferes with Hedgehog pathway), copper salts C12N 2500/20 (more specific symbols under C12N 2500/00 take precedence over C12N 2501/00). Where pathways intersect or overlap, precedence is given to the most specific symbol and multiple classification may well be considered. Head symbols (C12N 2501/10, C12N 2501/20, C12N 2501/30, etc.) should be used only for specific agents not (yet) foreseen in the detailed scheme. C12N 2501/998 - C12N 2501/999 serve as repository for proteins and chemicals which do not fit (yet) within the scheme. C12N 2501/50 and sub-symbols, are not intended to code for markers used in purification and/or identification of cells. (These are intrinsic properties of the cells, not reagents.)
Complements the C12N 2500/00 and C12N 2501/00 series to indicate conditioned media or co-culture conditions. Also used to index the components of artificial constructs and tissue equivalents: see C12N 5/0697.
Symbols for "remarkable" differentiation processes, i.e.:
- differentiation from one lineage to a different one, "lineages" being understood as the three dot hierarchies under C12N 5/0602, (i.e. going from C12N 5/0603 - C12N 5/0693).
- differentiation of pluripotent cells (ES C12N 5/0606, EG C12N 5/0611, iPS C12N 5/0696, multipotent adult stem cells C12N 5/0607)
- and also dedifferentiation, i.e. going backwards from a differentiated cell type to the corresponding stem/progenitor.
"Typical" differentiation processes from a lineage-specific stem/progenitor cell to its regular progeny within the same lineage should not be indexed.
C12N 2506/00 is occasionally used to index files pertaining to "rejuvenation" without any actual, specific and characterized/type-able resulting product (see C12N 5/16).
Used to spot the use of specific enzymes to digest tissues, i.e. not regular dispase/collagenase, or the use of very precise conditions for digestion.
Introduced with the closure of C12N 5/10.
C12N 2517/02 and C12N 2517/04 pertain to isolated cells from a transgenic or cloned animal (A01K 67/00, C12N 15/00 with A01K); in most cases, such documents do not actually belong to C12N 5/00. C12N 2517/10 documents pertain to cultivation steps which belong to C12N 5/00 (e.g. synchronisation of cells for nuclear transfer, maturation of oocytes for fecundation), although the ultimate purpose is still outside of C12N 5/00 and the document should be circulated accordingly.
Used in combination with C12N 5/06 (use of microsupports with a specific cell type).
Used in combination with C12N 5/0068 (mostly), but also C12N 5/0012 and C12N 5/06. Codes may be given either for the base material of the support or for coatings on said support.
Combination Sets (C-Sets):
In this subclass, C-Sets classification is applied to the following groups, listed in the table below, if the document discloses a pertinent combination of technical features that cannot be covered by the allocation of a single symbol. The fourth column of the table indicates the place where the detailed information about the C-Sets construction and the associated syntax rules can be found, in the definition section "Special rules of classification".
C-SETS ID | BASE SYMBOL | SUBSEQUENT SYMBOLS | C-SETS FORMULA; LOCATION OF C-SETS RULES |
#C12Na | (C12N, C12Q), DNA or RNA isolation/preparation process and its essential technical features; See C12N 15/10 | ||
#C12Nb | (C12N, C12Q), method for preparing vectors and its essential technical features; See C12N 15/64 | ||
#C12Nc | (C12N, C12N) Structure of a nucleic acid; see C12N 2310/00 |
The specific C-Sets rule is located at only one place of the base symbol in the section "Special rules of classification" in the definition. If the C-Sets rule is applicable to all groups of a subclass, it is located at the subclass level only. If the same C-Sets rule is applicable to multiple groups or subgroups within the same subclass, the C-Sets rule is placed at the highest group or subgroup of the multiple groups.
In this place, the following terms or expressions are used with the meaning indicated:
Antisense | DNA or RNA composed of the complementary sequence to the target DNA/RNA |
Aptamers | Oligonucleotide molecules that bind a specific target molecule. |
CpG-motifs | Cytosine-Phosphate-Guanine motifs; a cytosine is directly followed by a guanine in the DNA sequence; methylation of cytosine in CpG- motifs negatively regulates gene expression. |
Enzyme | Proteinaceous materials, which cause a chemical change in a starting material without being consumed in the reaction. |
Genetic Engineering | Technology used to alter the hereditary apparatus or gene structure of a living cell so that the cell can produce more or different chemicals, or perform completely new functions. |
Germ cell | Reproductive cells of the body, specifically, either egg or sperm cells. |
Maintaining | Supporting or sustaining growth or metabolic activity of microorganisms. |
Microorganism | Comprises single-celled organisms such as bacteria, actinomycetales or single-celled fungi, e.g. yeasts; for the purposes of classification, this term also includes viruses, human, animal or plant cells, protozoa, tissues and unicellular algae. |
Multipotent stem cell | A stem cell with the ability to give rise to multiple cell types belonging to one particular embryonic germ layer, the endoderm, the mesoderm or the ectoderm. |
Mutation | Any change that alters the sequence of bases along the DNA thereby changing the genetic material of a microorganism. |
NK cell | Natural killer cell |
Non-coding nucleic acid sequence | Nucleic acid sequence which does not contain instructions for making proteins. |
Pluripotent stem cell | A stem cell with the ability to differentiate into cells of at least two of the three embryonic germ layers, the endoderm, the mesoderm and the ectoderm. |
Preserving | Rendering microorganisms reversibly dormant. |
Proenzyme | An enzyme precursor |
Progenitor cell | A parent cell that gives rise to a distinct cell lineage by a series of cell divisions. |
Recombinant DNA Technology | Techniques for cutting apart and splicing together pieces of DNA from the same or different sources. |
Single-cell protein | Protein derived from microorganisms, usually bacteria or yeast, that are cultivated on a suitable medium and then harvested and processed for use as a food for livestock or humans. For example, blue-green bacterium Spirulina is processed and sold as a protein-rich health food. |
Stem cell | Cells capable of renewing themselves through mitotic cell division as well as differentiating into a diverse range of specialized cell types. The term covers adult stem cells as well as embryonic stem cells (ES) as derived from blastocysts. |
Totipotent stem cell | A stem cell with the ability to generate a whole organism autonomously; totipotent mammalian cells thus can differentiate into all three somatic lineages (endoderm or mesoderm or ectoderm), the germ line and extra embryonic tissues such as the placenta. |
Vector | A DNA sequence (e.g., plasmid, phage DNA) which may be employed to introduce a foreign gene into a host cell and is able to replicate autonomously in the host cell. |
This place covers:
Main group C12N 1/00 covers compositions comprising microorganisms, processes of treating microorganisms, and processes of culturing or growing of microorganisms.
Subgroups C12N 1/10, C12N 1/12, C12N 1/14, C12N 1/16, C12N 1/18, C12N 1/20 cover media compositions for a type of microorganism, compositions comprising a microorganism (with or without other compounds), processes of isolating, maintaining or propagating microorganisms which are specific for a class of microorganism.
Subgroups C12N 1/105, C12N 1/125, C12N 1/145, C12N 1/165, C12N 1/185, or C12N 1/205 covers a micro-organism per se which is a new natural isolate or a new mutant where the mutated gene is not known.
Subgroup C12N 1/04 covers methods of preserving or maintaining viable microorganisms, subgroups C12N 1/10, C12N 1/105, C12N 1/12, C12N 1/125, C12N 1/14, C12N 1/145, C12N 1/16 - C12N 1/185, C12N 1/20 and C12N 1/205 cover compositions of microorganisms irrespective of whether they are viable or not.
Subgroup C12N 1/36 also covers attenuation of pathogens for vaccine preparation.
This place does not cover:
Medicinal preparations containing material from microorganisms | |
Medicinal preparations containing material from algae | |
Medicinal preparations containing material from fungi | |
Preparing medicinal bacterial antigen or antibody compositions, e.g. bacterial vaccines |
Attention is drawn to the following places, which may be of interest for search:
Animal cells | |
Viruses | |
Enzymes | |
Carrier-bound or immobilised microorganisms | |
Treatment of microorganisms with electrical or wave energy | |
Preparation of mutants without insertion of genetic material, screening processes | |
Algae other than microalgae | |
Biocides, pest repellants or attractants, plant growth regulators containing microorganisms | |
Treating dough with microorganisms | |
Preservation of foods or foodstuffs, in general, using microorganisms | |
Fermentation of meat, fish | |
Preservation of meat, sausages, fish with microorganisms | |
Preservation of egg or egg products with microorganisms | |
Preservation of fruit or vegetables by acid fermentation | |
Preservation of edible seeds or cereals with microorganisms | |
Fermented milk preparations, treatment using microorganisms | |
Cream containing or treated by microorganisms | |
Butter preparation, addition of microorganisms | |
Butter having reduced fat content prepared by addition of microorganisms | |
Buttermilk containing or treated with microorganisms | |
Making cheese curd with microorganisms | |
Treating cheese curd after whey separation with microorganisms | |
Whey preparations containing microorganisms | |
Treating tea by fermentation with microorganisms | |
Tea extractions, extracts, with microorganisms | |
Coffee, removing unwanted substances, using microorganisms, preparations | |
Coffee, reducing or removing alkaloid content, using microorganisms, preparations | |
Extraction of coffee, coffee extracts, addition of or treatment with microorganisms | |
Treating cocoa by fermentation with microorganisms | |
Cocoa products containing microorganisms | |
Sweetmeats, confectionery or marzipan, containing microorganisms | |
Chewing gum containing microorganisms | |
Frozen sweets, e.g. ice confectionery, ice-cream, containing microorganisms | |
Obtaining protein compositions for feed-stuffs, from microorganisms | |
From cereals, wheat, bran, molasses by using microorganisms | |
From leguminous or other vegetable seeds from press-cake or oil-bearing seeds by treatment with microorganisms | |
From yeasts | |
Animal feeding-stuffs, addition of microorganisms | |
Preservation of green fodder by ensilage using microorganisms | |
Non-alcoholic beverages, fermented | |
Non-alcoholic beverages, clarifying or fining using microorganisms | |
Food or foodstuffs, fermentation of farinaceous cereal or cereal material, addition of microorganisms | |
Food or foodstuffs, treatment of pulse, fermented pulses or legumes, removing undesirable substances, using microorganisms | |
Food or foodstuffs, treatment of pulse, fermented pulses or legumes, addition of microorganisms | |
Meat products, meat meal, addition of or treatment with microorganisms | |
Meat products, meat meal, with yeasts or fungi | |
Meat products, meat meal, tenderised or flavoured meat pieces, addition of microorganisms | |
Egg products, addition of or treatment with microorganisms | |
Food-from-the-sea products, fish, fish meal products, addition of or treatment with microorganisms | |
Food consisting or nut meats or seeds, addition of or treatment with microorganisms | |
Food or foodstuffs containing microorganisms | |
Food or foodstuffs, modifying nutritive quality, containing bacteria or derivatives, e.g. probiotics | |
Food or foodstuffs, modifying nutritive quality, containing yeasts or derivatives | |
Tobacco products or substitutes, treatment with microorganisms | |
Processes for making harmful chemical substances harmless or less harmful by biological methods using microorganisms | |
Bioremediation, reclamation of contaminated soil microbiologically | |
Biological treatment of water, wastewater or sewage | |
Polypeptides, genes for polypeptides | |
Compositions for enhanced recovery of hydrocarbons, using bacteria (microbial enhanced oil recovery) | |
Refining of hydrocarbon oils by using microorganisms | |
Production of fats, pretreatment of raw materials by microorganisms | |
Detergent compositions containing microorganisms | |
Brewing of beer | |
Wine, other alcoholic beverages, preparation thereof | |
Vinegar, preparation thereof | |
Apparatus for enzymology or microbiology | |
Fermentation and enzymatic processes for the preparation of compounds and compositions; including processes involving microorganisms of different genera in the same process, simultaneously | |
Measuring or testing processes involving microorganisms | |
Processes using microorganisms | |
Enzymes | |
Purification of sugar juices using microorganisms | |
Extraction of sucrose from molasses using microorganisms | |
Extraction of metal components from ores or concentrates with the aid of microorganisms | |
Obtaining uranium using microorganisms | |
Directed molecular evolution of RNA, DNA, libraries, creating and screening processes (combinatorial chemistry) | |
Libraries contained in or displayed by microorganisms | |
Creation using whole viable microorganisms | |
Libraries contained in or displayed by microorganisms | |
Methods of creating libraries using whole viable microorganisms | |
Treating fibres, threads, fabrics with microorganisms | |
Processes for obtaining cellulose by treatment of cellulose-containing material (waste paper, pulp) with microorganisms | |
Treatment of cellulose-containing material with microorganisms | |
Pulp or paper, addition of microorganisms thereto | |
Pulp, addition of microorganisms thereto | |
Investigating or analysing water using microorganisms | |
Immunoassay, biospecific binding assay, for microorganisms | |
Biochemical fuel cells in which microorganisms function as catalysts | |
Biofuels |
The last place rule is applicable, but all information should be given a group. For example, if both yeast and bacterial compositions are part of the invention in the application, then classify in both (C12N 1/16 or C12N 1/165) and (C12N 1/20 or C12N 1/205).
In this place, the following terms or expressions are used with the meaning indicated:
Microorganism | Includes bacteria and other generally unicellular organisms with dimensions beneath the limits of vision which can be propagated and manipulated in a laboratory, including unicellular fungi (including yeasts), microalgae, protozoa. |
This place covers:
Methods of preserving or maintaining viable microorganisms which include protozoa, unicellular algae, fungi, bacteria, archaea, spores, viruses or bacteriophages.
Subgroups C12N 1/10 - C12N 1/205 cover compositions of microorganisms irrespective of whether they are viable or not.
Natural isolates are classified in C12R.
Preservation of excised body parts of humans or animals is classified in A01N 1/10.
This place does not cover:
Immobilized microorganisms |
This place covers:
Media compositions for protozoa, compositions comprising protozoa (with or without other compounds), processes of isolating, maintaining or propagating protozoa.
Attention is drawn to the following places, which may be of interest for search:
Microorganisms expressing a defined gene (if an operon is expressed) | |
Mutant microorganisms in which the mutation is defined (according to the defined mutation) | |
Microorganisms expressing a defined gene (according to the defined gene) | |
Processes for the preparation of a compound or substance using a microorganism |
Naturally isolated protozoa or mutant protozoa in which the mutation is undefined or cannot be defined are covered by C12N 1/105.
This place covers:
Media compositions for unicellular algae, compositions comprising unicellular microalgae (with or without other compounds), processes of isolating, maintaining or propagating unicellular microalgae.
This place does not cover:
Algae as new plants |
Attention is drawn to the following places, which may be of interest for search:
Naturally isolated microorganisms, or mutant microorganisms in which the mutation is undefined or cannot be defined, if an adapted or attenuated microorganism is obtained | |
Microorganisms expressing a defined gene (if an operon is expressed) | |
Mutant microorganisms in which the mutation is defined (according to the defined mutation) | |
Microorganisms expressing a defined gene (according to the defined gene) | |
Processes for the preparation of a compound or substance using a microorganism |
Looping references between C12N 1/12 and A01H 13/00 have been identified. Until this inconsistency is resolved in IPC, the current classification practice in CPC is as follows:
The reference should stay in the title of CPC until the correction/change is made in IPC.
Naturally isolated unicellular algae or mutant unicellular algae in which the mutation is undefined or cannot be defined are covered by C12N 1/125.
This place covers:
This place does not cover:
Culture of Mushrooms | |
Fungi as new plants |
Attention is drawn to the following places, which may be of interest for search:
Naturally isolated micro-organisms, or mutant micro-organisms in which the mutation is undefined or cannot be defined, if an adapted or attenuated micro-organism is obtained | |
Micro-organisms expressing a defined gene (if an operon is expressed) | |
Cosmetic or medicinal compositions or preparations comprising a micro-organism | |
Mutant micro-organisms in which the mutation is defined (according to the defined mutation) | |
Micro-organisms expressing a defined gene (according to the defined gene) | |
Processes for the preparation of a compound or substance using a micro-organism |
Looping references between C12N 1/14 and A01H 15/00 have been identified. Until this inconsistency is resolved in IPC, the current classification practice in CPC is as follows:
The reference should stay in the title of CPC until the correction/change is made in IPC.
Naturally isolated fungi, yeast or Saccharomyces, or mutant fungi, yeast or Saccharomyces in which the mutation is undefined or cannot be defined are covered by C12N 1/145, C12N 1/165 and C12N 1/185 and the appropriate C12R subgroup.
If the invention pertains to the attenuation of fungi, classification in C12N 1/36 should also be considered.
This place covers:
Media compositions for bacteria, compositions comprising bacteria (with or without other compounds), processes of isolating, maintaining or propagating bacteria.
Attention is drawn to the following places, which may be of interest for search:
Microorganisms expressing a defined gene (if an operon is expressed) | |
Mutant microorganisms in which the mutation is defined (according to the defined mutation) | |
Microorganisms expressing a defined gene (according to the defined gene) | |
Processes for the preparation of a compound or substance using a microorganism |
Naturally isolated bacteria, or mutant bacteria in which the mutation is undefined or cannot be defined are covered by C12N 1/205 and the appropriate C12R subgroup.
This place covers:
Processes for spore formation and for isolation of spores.
This place does not cover:
Attention is drawn to the following places, which may be of interest for search:
Processes using microorganisms |
In this place, the following terms or expressions are used with the meaning indicated:
Spore | Reproductive structure that is adapted for dispersal and surviving for extended periods of time, e.g. in unfavorable conditions. Spores form part of the life cycles of many microorganisms such as bacteria, microalgae, fungi and some protozoa. |
In patent documents, the following abbreviations are often used:
Spore | Endospore, exospore, myxospore, cyst |
This place covers:
- Animal cells, animal cell culture, differentiation in vitro are to be classified in C12N 5/00, provided that such products and processes are characterized and exemplified in the application. A cell which is merely claimed without real and exemplified provision thereof shall not be classified in C12N 5/00. This is the case - for example - where the examples only relate to differentiation of a stem cell to pancreatic cells, but where differentiation to hepatocytes or nerve cells is claimed. Then, only the differentiated pancreatic cells are classified. The other cells are not classified at all.
- Orthogonal Indexing Codes C12N 2500/00 - C12N 2539/10 are compulsory when classifying in this place. This relates to any specific and exemplified component of cell culture medium, active agent used in cell culture process, co-culture with or conditioned medium produced by, use of differentiation from one lineage to another or from pluripotent cells, use of cells in diagnostic, methods for the dissociation of cells, genetically modified cells, cells related to new breeds of animals, culture process characterised by the use of forces and support or coating for cells culture characterised by material, topography, treatment or properties. Any such additional information which is claimed but not exemplified is however not to be classified.
- Cells or tissues in medicinal preparations or for medical uses are classified in A61K 35/12 - A61K 35/65 irrespective of genetic modification or culture steps. Cells that are medically used after a relevant in vitro culturing step are classified in A61K 35/00 and C12N 5/00.
Further details of subgroups
The highest rank head group is to be used only in desperate cases, for documents which do not fit elsewhere, e.g. culture processes characterised by temperature
By exception to the dividing line general C12N 5/00 to the specific C12N 5/06, documents describing the modification of membranes of a specific cell type are classified both in C12N 5/0006 and C12N 5/06 and subgroups.
Only generic encapsulation here: Encapsulation of a specific cell type is C12N 5/06 (and subgroups), C12N 2533/00 Orthogonal Indexing codes may be used for the capsule material. Classifying the document in A61L 27/00 and/or A61K 47/00 should be considered. Encapsulation in fermenters is C12N 13/00. Note that encapsulated hepatocytes and pancreatic cells have specific subgroups: C12N 5/0671 and C12N 5/0677.
Generic media for potentially any type of cells. Culture processes using a particular agent are treated as a culture medium containing this agent (e.g. use of propranolol in cell culture: C12N 5/0018, C12N 2501/81). All documents should have at least one Orthogonal Indexing code from the range C12N 2500/00 - C12N 2502/00, and possibly many of them. Examples using CHO, 3T3, BHK or other common cell types should be regarded as generic disclosure and classified under C12N 5/00 rather than under C12N 5/06.
When a new medium is fully disclosed, most if not all C12N 2500/00 Orthogonal Indexing codes potentially apply; codes should be given for any useful information, i.e. any specific component with a specific concentration range (e.g. Ala 20 µ, Pro 10 µ … is indexed C12N 2500/32; reference to a standard composition of essential amino-acids as one component need not be indexed)—which can mean that many codes are actually given, but this appears to be the best way to retrieve reasonably quickly a document with a medium comprising, e.g., 10-100 nM Cu2+ or 20-75 µ Glu. It may not be necessary to index the most common additives (e.g. glutamine) and no code is foreseen for antibiotics, as these appear to be invariably present.
Note that serum-free media for specific cell types are C12N 5/06 (and subgroups), C12N 2500/99.
By "negative selection", it is meant that the document defines what is to be rejected but does not define in any positive manner what is to be retained. The combination of negative and positive markers is regarded as positive selection, and should normally allow to assign a specific type to the cells, and to classify in C12N 5/06 (and subgroups). In practice, C12N 5/0087 and C12N 5/0093 deal with purging blood or bone marrow (but also, rarely, of other tissues) from, respectively, immune cells and tumour cells before transplantation; C12N 5/0081 is thus very rarely used.
Documents should have at least one C12N 2533/00 Orthogonal Indexing Code. Supports for specific cell types are C12N 5/06 (and subgroups), C12N 2531/00 - C12N 2539/00.
Classification is made in the appropriate group below C12N 5/06; group C12N 5/06 implies that the teachings apply only to specific cells and it should always be possible to know whether the cells derive from vertebrates or invertebrates.
General Considerations:
The scope of any sub-group comprises the cells of the title, any tissue explant essentially consisting of these cells (e.g. skin = keratinocytes = C12N 5/0629), processes for isolating these cells (e.g. the definition of a set of markers to isolate a specific cell by FACS, but a document dealing with a specific antibody for one single marker can be classified solely in C07K 16/00), processes for preparing these cells (culture, differentiation), culture media and/supports specifically adapted for these cells and medical uses of these cells (as far as the nature of the cells is known and is relevant; e.g. medical composition of cultivated pancreatic cells C12N 5/0676, A61K 35/12, "low-tech" composition of animal tissue extracts A61K 35/00 and subgroups, photochemically-treated blood A61K 41/0057, A61K 35/14).
Controlled language: Title note of C12N 5/06: In this group, the following words are used with the meanings indicated:
- a "totipotent" cell can differentiate into all somatic lineages (ectoderm, mesoderm, endoderm), the germ line and extra embryonic tissues such as the placenta;
- a "pluripotent" cell is a somatic stem cell which can differentiate into cells of at least two of the three somatic lineages (ectoderm, mesoderm, endoderm);
- a "multipotent" cell is restricted to one lineage.
- "Progenitor" and "precursor" cells are further restricted within the lineage. If not explicitly foreseen, totipotent cells are classified with pluripotent cells. Multipotent cells should not be classified with pluripotent cells.
This has been adopted to clarify classification practice with respect to the use of the words "pluri-" and "multipotent." Under this definition, totipotent cells need to be able to generate placenta and amnion, i.e. only the zygote, blastula and morula cells strictly qualify; applicants tend to use the term more liberally … Conversely, the archetypal example of an adult pluripotent cell in the sense of C12N 5/0607, has been named "Multipotent Adult Stem (Progenitor) Cell" (MASC, MAPC) by its discoverers, which is too restrictive in view of our definitions.
Immediate precursors:
"Four dot" stem/precursor/progenitor groups usually cover (multipotent) stem cells as well as (restricted) precursors and committed progenitors, but there are two important exceptions with respect to the latter: myoblasts go with myotubes in C12N 5/0658 (since myotubes do not proliferate, culture can only be directed to their precursors). In the blood/immune hierarchy C12N 5/0634, immediate, committed, precursors are classified with their progeny; C12N 5/0647 is reserved for stem cells and multi-lineage progenitors.
Sub-headgroups C12N 5/0603, C12N 5/0608, C12N 5/0613, C12N 5/0618, etc vs. specific subgroups C12N 5/0604, C12N 5/0605, C12N 5/0606, C12N 5/0608:
The "three dot" (sub)headgroups are used for two different purposes:
- for cells whose precise type is not (yet) foreseen in the scheme,
- as a broad indication of end product where the examples do not provide enough evidence as to the result of a differentiation process
E.g. a document pertaining to neural differentiation of BM-MSC is C12N 5/0618, C12N 2506/1353 while a document pertaining to neuronal differentiation of BM- MSC, with detailed, extensive, evidence as to the neuronal phenotype is C12N 5/0619, C12N 2506/1353. In the intermediate situation where a document claims neuronal differentiation with only limited evidence (e.g. one or two markers), it is preferred to use the headgroup C12N 5/0618, especially if the document additionally provides examples of oligodendrocyte/glial differentiation at the same poor level of evidence.
The head group should be used only for specific vertebrate cells which do not fit elsewhere in the scheme. Generic disclosure for vertebrate cells is C12N 5/00 (and subgroups).
This is actually "embryonic or foetal cells and tissues", but interpreted in a very restricted sense to encompass only cells that are solely embryonic and/or foetal; embryonic/foetal cells of recognisable type which are also present in an adult are classified as adult cells. "Rejuvenated" cells claimed to have been brought back to pluripotency or cells described by an applicant as "ES-like" do not qualify for C12N 5/0606: These cells do not actually originate from an embryo or foetus, see C12N 5/0607(A). In practice, C12N 5/0603 itself only contains embryoid bodies and cells being in an intermediate stage of differentiation between pluripotent cells (ES) and "adult" (typable) tissue; e.g. "definitive endoderm" cells C12N 5/0603 (exists only in embryo), pancreatic cells C12N 5/0676 (adult). Embryonic germ (EG) cells, which can be regarded as an equivalent for ES, are C12N 5/0611, but embryonic carcinoma (EC) cells are classified in C12N 5/0606 as far as they are used as a model for ES (now that human ES have been isolated, EC technology appears to be obsolete).
Pluripotent adult stem cells are still a controversial and largely speculative topic. Extreme care must be exercised when allocating this class: to qualify, a cell must have demonstrated pluripotency (note that the corresponding examples of differentiation into at least two distinct lineages are not to be indexed) and must not belong to any other subgroup of the scheme.
This covers all kinds of (demonstrated) "rejuvenated" cells or induced pluripotent stem cells (iPS). The rejuvenation method/agents are to classified appropriately (C12N 2510/00 and C12N 2501/60 for typical iPS obtained by forced expression of Oct-3/4, Sox-2, Klf4, Nanog, cMyc…; C12N 2501/00, C12N 2500/00, C12N 2502/00 as appropriate for chemical agents). Detailed aspects regarding the construction of a suitable expression vector are to be classified in C12N 15/00.
The head group is to remain empty until a third mammalian sex is discovered or engineered. The few documents dealing both with oocytes and spermatozoa are classified in both subgroups C12N 5/0609 and C12N 5/061.
For practical reasons, precursors (myoblasts) have been grouped with their progeny (myotubes); as a result, only few documents remain in C12N 5/0659 (satellite cells).
Owing to the complexity of the lineage, committed precursors are grouped with their immediate progeny: e.g. pre-T cells go with T cells in C12N 5/0636; lymphoid stem cells, which can give rise either to B or T, remain in C12N 5/0647.
Vaccines: Immunogenic preparations (e.g. stimulated T cells, antigen-loaded dendritic cells or microphages) are classified in A61K 39/00 and subgroups according to the antigen with the appropriate A61K 2039/51 - A61K 2039/892 Indexing Code(s).
Immunotherapy: Cells classified in C12N 5/0634 - C12N 5/0639 are additionally classified in A61K 40/00 - A61K 40/50 with Orthogonal indexing codes A61K 2239/00 - A61K 2239/59, when the cells are modified for immunotherapy (e.g. CAR-T cells).
C12N 5/0629 contains purified keratinocytes and whole skin biopsies, assigned to what is regarded as the most relevant cell type, but artificially reconstructed skin is C12N 5/0698.
Encapsulation is assimilated to 3D culture. C12N 2533/00 Orthogonal indexing codes may apply to the capsule material.
For practical reasons, this group contains isolated vascular smooth muscle cells (not C12N 5/0661) as well as elaborated three-dimensional constructs comprising multiple types (e.g. vascular endothelium C12N 5/069, fibroblasts C12N 5/0656); in the latter case, the further cell types may be indexed with C12N 2502/00 Orthogonal Indexing codes as in C12N 5/0697.
Haemangioblasts, which are precursors for both the haematopoietic and the vascular endothelial lineages, always get a double class: C12N 5/0647, C12N 5/0692.
Tumour cells considered for themselves (i.e. as tumours) are classified here, but tumour cells used as convenient immortalised equivalents/models of their untransformed counterparts are classified as normal cells: e.g. embryonic carcinoma C12N 5/0606; medium specially adapted for hepatomas C12N 5/067, C12N 2500/00, C12N 2501/00, C12N 2502/00, C12N 2503/00, C12N 2506/00, C12N 2509/00.
Myeloma cell lines for use in the making of hybridomas are directly classified in C12N 5/163, with their application, rather than as C12N 5/0694.
"Tissue equivalent" is to be construed broadly as any in vitro construct associating different cell types to achieve one function; the precise structure or function of an actual tissue need not be achieved but it must be an artificial construct, not an actual tissue explant (C12N 5/0602 and subgroups, according to the source or dominant cell type), and no single cell type must be responsible for the desired effect, as is the case for a typical co-culture. Examples: model of neuromuscular junction where a neuron commands a muscle cell C12N 5/0697, C12N 2502/081, C12N 2502/1335; testicular prosthesis consisting of chondrocytes (for shape) and of Sertoli cells (for hormonal function), C12N 5/0697, C12N 2502/1317, C12N 2502/24; but embryonic stem cells on a feeder layer of embryonic fibroblast C12N 5/0606, C12N 2502/13. C12N 2502/00 codes are used to index all cell types, including accessory ones (e.g. vascular cells or tumour cells added in some illustrated embodiments to study vascular or tumour growth inside a tissue).
Not to be used. Genetic engineering itself is classified in C12N 15/00; cells modified for a particular application (e.g. recombinant expression, promoter-reporter constructs for testing) are classified with the application (C07K 14/00 - C07K 14/16, G01N 33/00, C12Q 1/00). Classification in C12N 5/00 is done only if there is actual interest in the cell itself; e.g. transfecting pdx1 in MSC to yield insulin-secreting cells (assimilated to pancreatic delta cells, according to the sought therapeutic effect, even though full differentiation and full functionality may not be achieved) C12N 5/0676, C12N 2506/1353, C12N 2510/00.
Has only been used for extremely rare cases of generic fused cell technology applicable equally to animal, vegetal, fungal (yeast) or bacterial cells.
Fusion partners are not indexed. C12N 5/163 covers also cell lines to be used as fusion partners, but not specific hybridomas producing a specific antibody.
Some "rejuvenated" cells prepared by introducing "young" cytoplasm into "old" cells, or even by transferring "old" nuclei into enucleated "young" cells" have been assimilated to fused cells and classified here. See also Orthogonal Indexing code C12N 2506/00. Note that nuclear transfer in itself is C12N 15/873 and circulation is warranted. Proper cloning is not classified here.
Immunotherapy: Cells classified in C12N 5/0634 - C12N 5/0639 are additionally classified in A61K 40/00 - A61K 40/50 with indexing codes A61K 2239/00 - A61K 2239/59, when the cells are modified for immunotherapy (e.g. CAR-T cells).
This place does not cover:
Plants reproduction by tissue culture techniques |
Attention is drawn to the following places, which may be of interest for search:
Plants | |
Mutation or genetic engineering; Use of hosts therefore | |
Use of cells in vaccines or immunological preparations | |
Gene therapy | |
Recombinant expression of proteins | |
Apparatus for cell culture | |
Apparatus for cell purification | |
Screening and/or testing using cells |
Classification Policy
Considering that:
- Documentation is a search tool;
- Laundry lists of further possible agents or applications, wishes (mere "hope to succeed") and plans for future extensive research are of little interest;
- If it is useful to cite a formal X document, such wish lists can easily be retrieved by full text search while actual technical content cannot easily be extracted from patent databases, due to the overall poor quality of abstracts and the noise generated in full text mode by the laundry lists; classification is based, and solely based, on what has actually been done, i.e. classification is based on the actual examples and not on the claims.
All relevant aspects of the examples, including unclaimed aspects, are classified as allowed by the scheme at the most specific place; the wording of the claims, e.g. with respect to further applications or possible generalisation, need not be considered and shall not be classified in the absence of actual support in the examples. The "last place rule" is not used; multiple symbols may be given as needed.
Irrelevant aspects, which are not classified are those upstream and/or downstream the invention (or "contribution to the art"):
- Upstream. If the invention is directed to the growth or differentiation of cells, the process used to obtain and purify the starting cell material;
- Downstream. Further testing with or about the grown or differentiated cells, e.g. checking for stemness by causing cells to differentiate, assessing the function of the resulting cells, in vitro testing for pharmaceuticals, etc.
A document whose examples are solely directed to testing in cells is not to be classified at all in C12N 5/00 but only either in G01N 33/00 or C12Q 1/00 (if emphasis is on the testing itself) or in A61K 31/00 - A61K 38/00 (if the intent lies with in vivo therapy after pre-clinical tests).
Non-patent literature is not classified, owing to its sheer abundance and to the quality of dedicated databases.
Some Principles and Practices
- Generic vs. Specific: C12N 5/00 and subgroups are used for general aspects, e.g. "universal" culture media or supports, while subgroups of C12N 5/06 are used for cell-type specific aspects; thus, classification is generally under C12N 5/06, with the appropriate deep Indexing symbol C12N 5/00. Examples using common cell types such as 3T3, CHO, BHK ("eucaryotic equivalent of E. coli") are classified as generic.
- Stem vs. Differentiated Cells: Classification is based on the end product as available to the skilled person. Documents pertaining to the purification of stem cells are classified as the stem cells; possible examples illustrating the potency of these cells to differentiate are not classified. Documents pertaining to the differentiation of stem cells are classified as the end product in the lower subgroups of C12N 5/0602, with all applicable symbols C12N 2500/00 - C12N 2502/00 and possibly C12N 2506/00; no symbol is given for the starting material, unless the document also discloses new protocols for its purification and/or maintenance. Documents pertaining to in vivo differentiation of stem cells are classified as the available product, i.e. as (a composition of) stem cells C12N 5/0606, C12N 5/0611, C12N 5/0623, C12N 5/0647, C12N 5/0662 - C12N 5/0668 , C12N 5/0672, C12N 5/0678, C12N 5/068, C12N 5/0687, C12N 5/0689, C12N 5/0692, C12N 5/0695, with the relevant symbol A61K 2035/124 for in vivo Application; there are no symbols for the resulting differentiated cells.
- Modified Cells, Fused Cells: C12N 5/10 is closed and should not be used; relevant information not related to the genetic engineering itself can be classified in C12N 5/06 with an C12N 2510/00 symbol.
The technology classified in C12N 5/12 and subgroups (fused cells, e.g. hybridomas C12N 5/163, hybridomas for producing a specific antibody C07K 16/00) appears to be mostly obsolete and technically deprecated.
The last overhaul in C12N 5/00 (from 2002 onwards) was driven by the surge of stem cell-related applications. In order to both keep track of differentiation processes and maintain stem cell groups to a manageable size, C12N 2506/00 was introduced. It also appeared that, with paperless classification and online search, it would be better to move cell-type specific media and supports to the corresponding cell group; C12N 2500/00 - C12N 2502/00 and, later C12N 2531/00 - C12N 2533/00, were introduced and C12N 5/00 documents deeply indexed during reorganisation (C12N 2531/00 - C12N 2533/00 indexing is less thorough because it was actually performed after reorganisation of C12N 5/06 was complete). To avoid duplicating in whole the existing hierarchies for proteins (A61K 38/00, C07K 14/00), enzymes (C12N 9/00), antibodies (C07K 16/00) and chemicals (A61K 31/00, C07, C08, etc), C12N 2501/00 was organised by signalling pathway, so that a ligand, its receptor, antibodies thereof and any "small organic molecule" with agonist or antagonist activity could share one single symbol.
Further cell groups were introduced, with mnemonic letters wherever possible. It has been submitted that the older layers of classification are organised by cell type while the newer layers are often in terms of organs; this is correct, but, taking into account that each different organ is associated with only a limited number of specific cell types, there should be no ambiguity in practice.
Blood and bone marrow-related groups were completely remodelled for more specificity, giving an opportunity to get rid of the lymphoid/myeloid distinction which proved to be problematic (notably dendritic cells are heterogenous and may derive from both lineages). In order to restrict as much as possible the large group for haematopoietic stem cells, committed progenitors were reclassified with their progeny but this policy was not consistently considered elsewhere.
C12N 5/0607 (pluripotent adult stem cells) derives from a practical issue, which ultimately stems from our poor understanding of stem cells in general. While stem cells are all the rage, it must be kept in mind that actually very few cells have been proved to be stem: Haematopoietic stem cells have only been "purified" to a few percent of a population, and that's with mouse cells, not even in humans. Mesenchymal stem cells are currently sourced from various tissues (bone marrow, circulating blood, adipose tissue) which are assumed to yield similar or identical cells; it has also not been demonstrated whether such MSC populations actually contain true multipotent cells rather than a mixture of various uni- or bipotent progenitors. Evidence of true stemness, at clonal level, is only available for embryonic stem cells; unfortunately, it is also known that ES are a laboratory artefact—a wonderful artefact, but an artefact nevertheless. C12N 5/0607 was intended as a temporary fix, carefully monitored, to be eventually dispersed into existing groups or better redefined, taking advantage of further insights into "stemness".
And then induced pluripotent stem cells (iPS) appeared, and a specific group was quickly needed to cover this technology.
Classification strategy according to subject-matter
For the sake of completeness, classification should be considered in all technical fields where they may be of interest. Common targets for additional classification include:
- A61K 35/00 (chemically "undefined" pharmaceutical compositions, including cells)
- A61K 39/00 (vaccines/immunology)
- A61K 48/00 (gene therapy)
- A61M (medical devices)
- C12M (apparatuses for cell culture)
- C12N 1/00 (bacteria, fungi)
- C12N 7/00 (viruses, including cells for virus culture)
- C12N 15/11(RNAi etc.)
- C12N 15/85 (vectors, transgenic animals)
Cells per se, tissues per se: C12N 5/0602, with the exception of vaccines.
Cells for vaccines are allocated a symbol in A61K 39/00 and subgroups according to the antigen, a symbol in A61K 2039/51 and subgroups and a symbol in C12N 5/00 and subgroups, when the invention is directed to the culture process.
For cellular immunotherapy, a symbol can be allocated in C12N 5/0634 or C12N 5/0636 - C12N 5/064 with an appropriate symbol in A61K 40/00 and subgroups and A61K 2239/00 - A61K 2239/59 if the invention is the invention is directed to the culture process.
Medical preparations containing living animal cells (other than vaccines):
C12N 5/0601, C12N 5/0602, etc with the relevant symbol A61K 35/12 or A61K 48/00.
Considered for classification in A61K 35/00.
Transformed cells, Immortalised cells: Quite often, transfected cells are nothing more than a means to produce a protein (C07K 14/00, C07K 16/00, C12N 9/00, possibly gene therapy, A61K 48/00) or to perform biochemical tests or screening assays (G01N 33/50 or C12Q 1/02): these cases do not deserve classification in C12N 5/00 and no further C12N 2510/00 symbol is required. If, and only if, the document is classified in C12N 5/00 for another reason or the transformation itself is important (e.g. a cell immortalised by telomerase, C12N 2510/04), an C12N 2510/00 symbol is used in combination with the C12N 5/06 class of the cell type. If the cell type is not relevant, the document pertains to a general method of transfection and should be classified in C12N 15/00.
Also, claims to a transformed cell isolated from a transgenic animal (possibly made for that sole purpose) need not be classified in C12N 5/0602 even the cell type is specified: Consider classification in C12N 15/85 and add an additional C12N 2517/00 symbol. The same apply to fused cells (e.g. hybridomas), which are classified in C12N 5/12 and subgroups only if their interest goes beyond the antibody they produce (forward to C07K 16/00).
Apparatuses for cell purification are not classified at all in C12N 5/00; if the application concerns a particular (novel) reagent, a particular device or a method specifically tied to a particular device, in many cases it will be classified fully outside of C12N 5/00. The typical document retained in C12N 5/00 pertains to the use of a combination of (known) reagents (combinations of markers) for isolating a given cell type.
Culture medium:
C12N 5/0018 - C12N 5/0056 for a "universal" medium, including medium for "generic cells".
C12N 5/06 and subgroups for a medium dedicated to a specific cell type.
In all cases, all specified components are classified with C12N 2500/00 - C12N 2502/00 symbols. Many documents pertaining to culture media fall under C12N 5/06 in CPC, but belong to C12N 5/00 and subgroups thereof under IPC.
Culture support:
C12N 5/0068 and C12N 5/0075 possibly add a C12N 5/06 (and subgroups) class for the cell used in the examples if it is relevant. C12N 5/0601, C12N 5/0602… in the less common case where the support is type-specific classify with C12N 2533/00 and C12N 2531/00 symbols.
Cell culture apparatus:
C12M Outside of the scope of this document.
Cell culture process:
C12N 5/0601, C12N 5/0602 etc.
Classify active agents with C12N 2501/00, C12N 2500/00, C12N 2502/00 symbols as applicable.
Differentiation process:
C12N 5/0601, C12N 5/0602 etc. for the resulting cell.
Classify differentiation agents with C12N 2501/00, C12N 2500/00, C12N 2502/00 symbols as applicable.
The differentiation of a lineage-restricted stem cell into the corresponding, expected, progeny (from a C12N 5/0606, C12N 5/0611, C12N 5/0623 etc progenitor or stem cell to a C12N 5/0603, C12N 5/0608, C12N 5/0613 etc terminal cell within the same branch of the hierarchy) is not further classified; differentiation of pluripotent cells (C12N 5/0606, C12N 5/0607, C12N 5/0611) and "unexpected" differentiation processes, such as transdifferentiation to a different branch of the hierarchy or dedifferentiation (from a differentiated cell into a more primitive or more potent cell), are classified with a C12N 2506/00 symbol for the initial cell.
Tissue culture, 3D culture, tissue equivalents:
C12N 5/0062 for a generic process.
C12N 5/0601, C12N 5/0602 etc. for a specific tissue defined by one single cell type or for ex vivo culture of tissues (e.g. skin explants C12N 5/0677).
C12N 5/0697 etc. for an artificial tissue construct requiring the association of more than one type of cells to achieve its function; classify all cell types with C12N 2502/00 symbols.
Tests, screening assays on cells or tissue equivalents:
Classify in G01N 33/50 or C12Q 1/02.
Classification with C12N 2503/00 symbols is only intended to avoid superfluous double classification when the cell in itself is of interest for C12N 5/00 and the testing process is not inventive.
See also A01H 4/00 through A01H 4/008
See also A01H 4/00 through A01H 4/008, C12N 1/12, C12N 5/14 and A01G 33/00
A "pluripotent" cell is a somatic stem cell which can differentiate into cells of at least two of the three somatic lineages (ectoderm, mesoderm and endoderm). Totipotent cells are classified with pluripotent cells. A "totipotent" cell can differentiate into all somatic lineages (ectoderm, mesoderm and endoderm), the germ line and extra-embryonic tissues such as the placenta
A "pluripotent" cell is a somatic stem cell which can differentiate into cells of at least two of the three somatic lineages (ectoderm, mesoderm and endoderm). Totipotent cells are classified with pluripotent cells. A "totipotent" cell can differentiate into all somatic lineages (ectoderm, mesoderm and endoderm), the germ line and extra-embryonic tissues such as the placenta
A "multipotent" cell is restricted to one lineage.
Containers specially adapted for collecting and storing blood and plasma are classified in A61J 1/05.
This place does not cover:
Preservation of excised living parts of human bodies |
This place does not cover:
Preservation of excised living parts of human bodies |
Attention is drawn to the following places, which may be of interest for search:
Containers specially adapted for collecting, storing or administering blood or plasma |
This place covers:
Containers not comprising mechanical means that actively alter temperature. The use of phase change materials, e.g. ice, to passively keep the cells or tissues cool is not considered "active" refrigeration.
This place covers:
Documents specifically addressing a transport aspect, e.g. providing means to counteract shocks or tilting.
Containers that are suitable for transport but do not comprise provisions specifically for transporting should be classified in C12N 5/542, C12N 5/546 or C12N 5/547.
Processes comprising temperature changes inherently involved in freezing or cryopreservation wherein no specific technical effect is disclosed for the temperature profile are not classified in this group.
This place covers:
Viruses are ultramicroscopic (20 to 300 nm in diameter), metabolically inert, infectious agents that replicate only within the cells of living hosts, mainly bacteria, plants, and animals: composed of an RNA or DNA core, a protein coat, and, in more complex types, a surrounding envelope.
Note that a virus (or VLP) as such is not considered a nanoparticle even if it falls under the size constraints of nanotechnology (see B82Y 5/00).
- Viruses as such, e.g. new isolates, mutants or their genomic sequences
- New viral genes, new structural/functional aspects of known viral genes
- Virus like particles (VLP)
- Uses of virus other than therapeutic or vaccine, e.g. disinfectant
- Use of virus as therapeutic agent, other than vaccine, e.g. as cytolytic agent
- Use of viral protein as therapeutic agent other than vaccine, e.g. apoptosis inducing or anti-inflammatory
- Use of virus or viral component as vaccine, e.g. live-attenuated or inactivated virus, VLP, viral protein
- Use of virus or viral particle as a vehicle, e.g. encapsulating small organic molecule
- Use of virus or viral particle as a vector, e.g. encapsulating viral genome or elements thereof as genetic vector
- Chimeric viral vector comprising heterologous viral elements for production of another viral vector
- Special targeting system for viral vectors
- Methods of production or purification of viral material relating to complementing cells and packaging systems for producing virus or viral particles
- Methods of inactivation or attenuation by genetic engineering, by chemical treatment, by serial passage
- Demonstrated in vivo effect
This place does not cover:
Phage display | |
Therapeutic viral non-coding nucleic acid (siRNA) | |
Antiviral drugs | |
Antibodies against viral antigens | |
Diagnostics using viral genes | |
Diagnostics using viral protein |
Attention is drawn to the following places, which may be of interest for search:
Transgenic animal model for infectious disease | |
Further aspects of vectors | |
Animal model for infectious disease | |
Virus capsids or envelopes enclosing drugs | |
Therapeutic use of virus not as vaccine | |
Therapeutic use of viral proteins, not as vaccine | |
Further aspects of (viral) vaccines | A61K2039/00 |
Viral vaccine | |
Polyvalent vaccine | |
Vaccine characterised by adjuvant | |
Gene Therapy | |
Fusion proteins | |
Measuring or testing involving virus or bacteriophage |
The complexity of the field is compounded by the large number of available viruses and the increase in their corresponding (therapeutic) uses. In this scheme, taxonomic indexing codes are assigned (C12N 2710/00-C12N 2796/00 series to define which virus is used in a given invention. These codes are extended by two digits to define the specific use(s) of the virus or viral component.
Allocation of these codes is compulsory for all relevant virus subject-matter, according to the rules defined below.
The use of the subdivisions in C07K 14/005-C07K 14/19, C12N 7/00-C12N 7/08 and C12N 15/86-C12N 15/869 is discontinued since the taxonomy within said classification ranges is incomplete and inconsistent. Only A61K 39/12, C07K 14/005, C12N 7/00 and C12N 15/86 will be given in relevant cases, as indicated below.
C12N 7/00 should be given when no other viral (A61K 39/12, C07K 14/005, C12N 15/86) or other group is appropriate, since giving at least one group as invention information is mandatory.
The specific codes in the C12N 2710/00 - C12N 2796/00 ranges have the format C12N27xx/xxxyy. The first part of the code indicates the structural element of the invention, namely the specific virus to which the invention relates. The two last digits before the '/' and the first three digits after the '/' represent the taxonomic location of the virus and the last place rule applies:
- C12N2710/xxx: double stranded DNA virus
- C12N2720/xxx: double stranded RNA virus
- C12N2730/xxx: reverse transcribing DNA virus
- C12N2740/xxx: reverse transcribing RNA virus
- C12N2750/xxx: single stranded DNA virus
- C12N2760/xxx: single stranded RNA virus negative-sense
- C12N2770/xxx: single stranded RNA virus positive-sense
- C12N2780/xxx: viroids and subviral agents
- C12N2790/xxx: naked RNA Virus
- C12N2792/xxx: archaeabacteria virus
- C12N2795/xxx: bacteriophage
Specific further aspects of viruses that are sufficiently disclosed in the application and worth classifying are assigned in combination with the taxonomy by addition of codes of two digits (yy) at the end of the taxonomic information (xx/xxx). The scope of the information covered by the last two digits is as defined below.
If multiple functions are disclosed for the same virus or viral component, multiple codes of the format C12N27xx/xxxyy are given:
- 11: General: This code covers aspects of viruses that do not fall under the codes defined below. Codes 21-64 take precedence.
- 21: Viruses as such: This code corresponds to IPC C12N 7/00 and covers completely new virus species, as well as new isolates of known viruses. Said viruses may be for instance defined by their genomic sequences or deposit numbers. Disclosed uses of said viruses as indicated below are additionally classified. If the virus per se is an essential feature of the invention, C12N 7/00 is to be given.
- 22: New viral proteins or individual genes, new structural or functional aspects of known viral proteins or genes: This code corresponds to IPC C07K 14/005-19 and encompasses newly identified open reading frames and their expression products, as well as newly identified forms of known viral proteins and genes. Furthermore new mutants, fragments, epitopes and fusion proteins of viral proteins are included. When such gene or protein relates to self-assembling capsids, code 23 for VLP is given. Codes in the C07K 2319/00 range are to be added for disclosed fusion proteins. Additional aspects of said proteins and genes as listed below should be considered, as well as aspects covered by other parts of the classification scheme e.g. in diagnostics (G01N 33/00, C12Q 1/00) for antibody production (C07K 16/00), in therapy (A61K 38/162), with enzyme function (C12N 9/00), using RNAi for viral protein (C12N 15/11), codon optimised (C12N 2800/22). If the protein or its gene is an essential feature of the invention, also C07K 14/005 is to be given, together with the code ending with 22.
- 23: Virus like particles, VLP: VLPs to be classified here are self-assembling viral protein complexes lacking the corresponding viral genome. This code is only to be given when the VLP is an essential feature of the invention.
- 31: Uses of virus other than therapeutic or vaccine, e.g., use of bacteriophages as disinfectant: N.B. , 'Processes using viruses or cell lines' is not used.
- 32: Use of virus as therapeutic agent, other than vaccine, e.g. cytolytic viruses for tumor therapy: The class A61K 35/76 should be considered in accordance with the classification rules in said field, when the therapeutic activity is an essential feature of the invention.
- 33: Use of viral protein for therapeutic application other than vaccine: This code corresponds to IPC A61K 38/16, and is given for viral proteins that have therapeutic activity, for instance cytotoxic, apoptosis inducing or anti-inflammatory viral proteins. If the therapeutic use of the viral protein is an essential feature of the invention, also A61K 38/16 should be considered in accordance with the classification rules in said field.
- 34: Use of virus or viral component as vaccine: This code corresponds to IPC A61K 39/12-29 and covers the various types of viral vaccines, e.g. live attenuated or inactivated virus, VLP, viral protein or nucleic acid. Further classification of the vaccine aspects needs to be done in accordance with the specific classification rules in said field. In particular, if multiple viral antigens are combined of which at least one is viral, A61K 39/295 needs to be given; if the vaccine is characterised by the adjuvant, A61K 39/39 should be added. The relevant codes from A61K2039/00 for further vaccine aspects should also be given. If the use as a vaccine is an essential feature of the invention(s), also A61K 39/12 is to be given, together with the code ending with 34.
- 41: Use of virus or viral particle as a vector, specific codes 42-45 take precedence.
- 42: Use of viral particle as vehicle: This code is given when no genetic material derived from the viral genome is transported by the viral vector, but rather encapsulated agents, e.g. small nucleic acids or organic compounds. Codes in the C12N 2810/00 series needs to be considered when the surface of the vector is altered to influence targeting.
- 43: Use of viral genome or elements thereof as genetic vector: This code corresponds to IPC C12N 15/86-C12N 15/869 and is given when the invention resides in the vector, for instance when no such virus was previously known as genetic vector, when a known viral genetic vector is improved, or when a new property (e.g. tissue tropism) is uncovered and results in (a) new application(s) of a known viral vector. Additional codes from the C12N 2800/00-C12N 2840/00 ranges need to be considered. If the vector is an essential feature of the invention, also C12N 15/86 is to be given together with codes ending with 43-45.
- 44: Chimeric viral vector comprising heterologous viral elements for production of another viral vector: This code is reserved for the viral backbone of said vector, i.e. if a vaccinia viral vector is used to express a lentiviral vector system, the vaccinia aspect is indicated with 44 and the lentiviral vector with 43.
- 45: Special targeting system for viral vectors: Influencing cell type specificity by modification of the surface of the virus, the specific targeting element can be further defined by codes in the C12N 2810/00 series. If altered tropism is only effected by transcriptional or translational elements, give the relevant codes of the C12N 2830/00 or C12N 2840/00 series.
- 51: Methods of production or purification of viral material: This code corresponds to IPC C12N 7/02 and relates to new protocols for producing viral material, e.g. using particular cell types, growing conditions to increase production, or novel purification methods, e.g. defined by specific centrifugation or chromatography steps. Also methods of stabilising virus compositions using cryopreservants or other excipients are classified here. If production, purification or stabilisation are an essential feature of the invention, also C12N 7/00 is to be given, together with the code ending with 51.
- 52: relating to complementing cells and packaging systems for producing virus or viral particles: This code relates to packaging cells that stably or transiently express viral genes to allow production of virus lacking said genetic information on their genome. This code is also to be given if other aspects of packaging are an important aspect of the invention. If the cells or packaging systems are an essential feature of the invention, also C12N 7/00 is to be given, together with the code ending with 52.
- 61: Methods of inactivation or attenuation: Codes 62-64 take precedence, this code is only used for methods of inactivation or attenuation not covered below, such as (UV) irradiation. If inactivation or attenuation is an essential feature of the invention, also C12N 7/00 is to be given together with codes ending with 61-64.
- 62: Methods of inactivation or attenuation by genetic engineering: This code is given when the nature of the attenuating mutation is known and can be reproduced. Virus-like particles are not classified here but with code 23.
- 63: Methods of inactivation or attenuation by chemical treatment.
- 64: Methods of inactivation or attenuation by serial passage: Selection of new viral strains by (further) serial passage on cell lines, for instance taking modified vaccinia virus Ankara 575 and submitting it to further rounds of serial passage on cells. If mutations have been identified with proven relationship to attenuation, also 62 should be given.
- 71: Demonstrated in vivo effect: This code is given when credible in vivo data are presented that are indicative for a (therapeutic) effect in human or animals. For instance data from clinical trials or from highly relevant animal models, such as primates for HIV, or showing protection of cattle from viral disease, e.g. preventing vertical transmission.
This place covers:
- Enzymes or proenzymes and compositions containing enzymes or proenzymes.
- Processes for preparing, activating, inactivating, inhibiting, stabilizing, separating, or purifying enzymes.
- Genes and other polynucleotides coding for enzymes.
- Non-coding nucleic acid sequences, e.g. promoters, operators, derived from genes or operons coding for enzymes.
- Fragments of enzymes and nucleic acids encoding enzymes (fragments of less than 5 amino acids are also classified in C07K)
- Fusion proteins comprising an enzyme or part thereof.
- Antibodies with enzymatic/catalytic activity, e.g. abzymes.
- Crystallized enzymes.
- Biocidal, pest repellent, pest attractant or plant growth regulatory activity of compounds or preparations containing enzymes is further classified in subclass A01H.
- Therapeutic activity of compounds containing enzymes, is further classified in subclass A61K.
- Uses of cosmetics or similar toilet preparations containing microorganisms or enzymes are further classified in subclass A61Q.
- Fragments of less than 5 amino acids are also classified in maingroup C07K 5/00.
- Carrier-bound or immobilised enzymes and preparations thereof are further classified in maingroup C12N 11/00.
- Treatment of enzymes with electrical or wave energy is further classified in maingroup C12N 13/00.
- Operons comprising the genes for several enzymes are also classified in C12N 15/52.
- Fusion polypeptides comprising (part of) an enzyme are further classified in C07K 2319/00.
- Crystals of enzymes are also classified in C07K 2299/00.
This place does not cover:
Catalytic nucleic acids e,g, ribozymes | |
Antisense nucleotides against enzymes | |
Antibodies against enzymes | |
Measuring or testing processes involving enzymes; Compositions therefore (including kits, test papers etc); Preparing such compositions | |
Immunoassays for enzymes |
Examples of places where the subject matter of this place is covered when specially adapted, used for a particular purpose, or incorporated in a larger system:
Compositions containing enzymes and use of the compositions and compounds for preservation of bodies of humans or animals or parts thereof | |
Compositions containing enzymes and use of the compositions and compounds for preservation of plants or parts thereof | |
Biocides, pest repellents or attractants or plant growth regulators containing enzymes | |
Bakery products which may contain enzymes | |
Foods or foodstuffs containing enzymes | |
Body treating or pharmaceutical preparations containing enzymes | |
Medicinal preparations containing nucleic acids encoding enzymes | |
Medicinal preparations containing genetic material encoding enzymes which is inserted into cells of the living body to treat genetic diseases; Gene therapy | |
Bandages, dressings or absorbent pads for physiological fluids containing enzymes | |
Antibodies against enzymes | |
Enzyme containing detergent compositions | |
Preparation of malt | |
Preparation of compounds using enzymes | |
Measuring or testing processes involving enzymes; Compositions therefore (including complete kits, test papers etc); Preparing such compositions | |
Immunoassays for enzymes |
Attention is drawn to the following places, which may be of interest for search:
Preparing polynucleotides using enzymes | |
Food compositions | |
Chemical aspects of, or use of materials for, bandages, dressings, absorbent pads or surgical articles | |
Peptides having more than 20 amino acids being receptors (as they often possess also an enzymatic function) | |
Preparation of compounds by using enzymes |
Instead of the "last place rule" the following rules are applicable for C12N 9/00 and subgroups:
- Enzymes are generally categorised according to the "Nomenclature and Classification of Enzymes" (as valid on 1 April 2011) of the International Commission on Enzymes. Where appropriate, this designation appears in the subgroups in parenthesis. If the information in disclosures is not enough to be able to classify the document in a subgroup for one specific enzyme, classification is done in a higher, less specific, group. This implies that in case of a search for a specific enzyme also such higher groups should be consulted.
- Besides the enzyme itself also fragments (like epitopes) or derivatives (like mutants) thereof, its proenzyme or just its signal sequence, their encoding polynucleotides or non-coding parts (like promoters, operators) of its gene, including methods for their preparation are all classified in the same subgroup. (antisense DNA or RNA against enzymes C12N 15/1137)
- Documents are only classified in main group C12N 9/00 and lower if there is more disclosed of an enzyme than just a name, a supplier, a reference to a further document for its production and the like. Properties like a sequence or mutation, or a method for its production etc. must be disclosed.
- Documents disclosing only lists of sequences with (putative) enzymatic activity are only classified in the last common enzyme group without classes for each mentioned enzyme, unless more information is given than just the sequence with a putative function. For example a long list of sequences comprising only (putative) proteases will be classified in C12N 9/48. If there are also lipases disclosed the general class for hydrolases (C12N 9/14) will be given. A combination with an acyltransferase results in C12N 9/00.
- Documents relating to/being valid for only one specific enzyme, like a mutant of a known enzyme, a new enzyme or a purification method specifically adapted to a particular enzyme, are only classified in the most specific and subgroup relating to this specific enzyme. However, it can also be clear from the provided information that, although the examples concern only one enzyme, the invention is also valid for other enzymes (like a specific purification method only exemplified for a pentosyltransferase but applicable for all other glycosyltransferases). Such documents are both classified in the most specific and subgroup for the enzyme in the examples and in a higher common group (in this case C12N 9/1048).
- C12N 9/00 (and C07K 14/00) vs. C12N 15/00: C12N 9/00 (and C07K 14/00) stop where C12N 15/00 begins. C12N 9/00 (and C07K 14/00) are only used for the product (the inventive non-coding sequence of a gene) while C12N 15/00 is used for the use of this product (e.g. a promoter present in a vector for the production of other proteins). Non-coding sequences are only classified in C12N 9/00 (or C07K 14/00) if they are (part of) the invention. If the non-coding sequence is just an arbitrary choice from more available sequences it is not classified in C12N 9/00 (or C07K 14/00)
- Reporter genes/enzymes like alkaline phosphatase, beta-lactamase etc. are not classified if they are just present for the detection of something else irrespective whether the above prerequisites of sequence, being prepared and the like are fulfilled. If the document however concerns reporter genes with a specifically mentioned property or preparation, classification in the proper group is necessary (fusion polypeptides containing such reporter enzymes are also classified in C07K 2319/61)
- Proteins with more than one function, like many receptors which often also have an enzymatic function, or the so called oncogenes, which, besides being tumour related, also have their original function, are classified in all groups concerning their different functions/activities. Depending on the discovery of further different functions older documents may still be present in only one class. Such documents are not systematically reclassified. For search purposes both groups should be consulted.
- Enzymes prepared by recombinant DNA technology are not classified according to the host used, but according to the original organism from which the encoding nucleotides were obtained, e.g. HIV protease expressed in E. coli is classified with viral proteases
- Rules for C12Y in addition to C12N 9/00: Enzymes are classified additionally in C12Y according to their full EC number (EC a.b.c.d.). An additional C12Y symbol is also allocated even when a subgroup specific for a single enzyme (EC number) already exists in C12N 9/00. Thus, thrombin (EC 3.4.21.5), for which a single subgroup, C12N 9/6429, exists, will additionally be allocated the appropriate C12Y symbol. (This double classification is of potential use for other areas of technology (e.g. food chemistry, detergents) where C12Y classification is used because a more specific classification scheme for enzymes in these areas is not present). A less specific C12Y symbol (e.g. EC a.b.c.) is only used in addition to a C12N 9/00 symbol if the subject-matter to be classified concerns a group of enzymes with a common activity AND there is no equivalent group present in C12N 9/00 for enzymes with the common activity. For example, a document describes the purification of carboxy-lyases (EC 4.1.1.). C12N 9/88 (lyases, EC 4.) is the closest symbol in C12N 9/00, thus the document is classified in C12N 9/88 and C12Y 401/01. However a document describing the purification of hexosyl transferases is classified ONLY in C12N 9/1051.
In this place, the following terms or expressions are used with the meaning indicated:
Antisense | DNA or RNA composed of the complementary sequence to the target DNA/RNA |
Enzyme | Proteinaceous materials, which cause a chemical change in a starting material without being consumed in the reaction. |
Fusion polypeptide | A polypeptide consisting of (parts of) two or more different proteins covalently linked to each other by a peptide bond |
Kit | A collection of individual reagents for use in an assay grouped together but being present as separate entities in separate compartments |
Mutation | Any change that alters the sequence of bases along the DNA thereby changing the genetic material of a microorganism. |
Non-coding nucleic acid sequence | Nucleic acid sequence which does not contain information for the amino acid sequence of a gene product. |
Operon | A DNA construct containing a cluster of genes under the control of a single regulatory signal or promoter |
Proenzyme | An enzyme precursor |
Ribozyme | RNA molecule capable of catalysing a chemical reaction |
Signal sequence | A 3-60 amino acids long peptide that directs the transport of the protein that is attached to it |
This place covers:
Proteinases (, e.g. Endopeptidases (3.4.21 - 3.4.25)) derived from bacteria or proteinases derived from Archaea, formerly known as Archaebacteria.
This place covers:
- General methods for the stabilisation of enzymes.
- Adducts of enzymes with a specific compound resulting in a more stabile enzymes and their preparation.
- Enzyme compositions with specific physical properties of pH, temperature, concentration etc. resulting in a better stability of the enzymes and their preparation.
- Enzyme compositions comprising additives resulting in a better stability of the enzymes and their preparation.
- Enzyme conjugates being more stabile than the unconjugated enzyme and their preparation.
Carrier-bound or immobilized enzymes are also classified in C12N 11/00.
In this place, the following terms or expressions are used with the meaning indicated:
Stabilisation | preserving activity in time and/or amount under certain conditions of pH, temperature etc. |
Adduct | a product of a direct addition of two or more distinct molecules, resulting in a single reaction product containing all atoms of all components |
Enzyme conjugate | enzyme covalently bound to one or more disctinct molecules |
This place does not cover:
Preparation of granular or free-flowing enzyme compositions if it is done to stabilise the enzyme |
This place covers:
Enzymes or microbial cells that are immobilised or bound to a carrier, and processes for the immobilisation or binding to carriers of enzymes and microbial cells, with the aim of using them as immobilised or carrier-bound enzymes or microbial cells.
This place does not cover:
Immobilised or carrier-bound peptides or proteins that are not enzymes | |
Processes and methods wherein immobilised or carrier-bound enzymes or microbial cells are used, e.g. fermentative or enzymatic processes for the preparation of compounds or compositions | |
Processes and methods wherein immobilised or carrier-bound enzymes or microbial cells are used, e.g. measuring or testing methods involving immobilised or carrier-bound enzymes |
Attention is drawn to the following places, which may be of interest for search:
Microorganisms, compositions thereof | |
Undifferentiated human, animal or plant cells | |
Enzymes | |
New plants or processes for obtaining them; plant reproduction by tissue culture techniques | |
Biological treatment of water, waste water or sewage | |
Apparatus for the use of immobilised or carrier-bound enzymes | |
Processes using microorganisms | |
Enzymes |
The last place rule is applicable, but all information should be given a class. E.g. if a bridging agent is used and the carrier is carbohydrate and both are part of the invention in the application, then classify in both C12N 11/06 and C12N 11/10.
In this place, the following terms or expressions are used with the meaning indicated:
Microbial cell | Microorganisms, including bacteria and other generally unicellular organisms with dimensions beneath the limits of vision which can be propagated and manipulated in a laboratory, including unicellular fungi (including yeasts), microalgae, protozoa and, moreover, human, animal and plant cells. |
This place covers:
Processes of treating microorganisms or enzymes with electrical or wave energy including magnetic and sound waves.
Attention is drawn to the following places, which may be of interest for search:
Microorganisms, compositions thereof | |
Undifferentiated human, animal or plant cells | |
Enzymes | |
Introduction of foreign genetic material using processes not otherwise provided for | |
New plants or processes for obtaining them; plant reproduction by tissue culture techniques | |
Gene therapy | |
Means for application of stress for stimulating the growth of microorganisms or the generation of fermentation or metabolic products | |
Processes using microorganisms | |
Enzymes |
In this place, the following terms or expressions are used with the meaning indicated:
Microorganism | Bacteria and other generally unicellular organisms with dimensions beneath the limits of vision which can be propagated and manipulated in a laboratory, including viruses and unicellular fungi (including yeasts), algae (including microalgae) and protozoa and, moreover, human, animal and plant cells. |
This place covers:
Preparing mutants and screening processes therefor.
Processes of fusing two or more cells to each other.
Recombinant DNA-technology including:
- Processes for manipulating genetic material;
- Processes of preparing, isolating and purifying nucleic acids;
- Methods for the introduction of genetic material into cells using vectors or other expression systems, using microencapsulation, using microinjection, and other ways;
- Methods of regulating gene expression;
- Non-coding nucleic acid sequences, e.g. Promoters, operators, enhancers, suppressors, silencers, locus control regions, antisense nucleic acids, and aptamers, used in regulating gene expression or in other recombinant DNA technology related methods. (Non-coding parts of genes are also classified in the subgroup corresponding to the product of the gene in C07K 14/00 or C12N 9/00, only if they are part of the invention).
- Operons
- Processes for the preparation of fusion proteins
This place does not cover:
Nucleic acids not used in recombinant technology and their chemical preparation | |
Genes and other polynucleotides coding for peptides per se |
Attention is drawn to the following places, which may be of interest for search:
Classification in C12N 15/00 should only be performed in exceptional cases in the absence of a more specific subgroup.
C12N 9/00 (and C07K 14/00) vs. C12N 15/00: C12N 9/00 (and C07K 14/00) stop where C12N 15/00 begins. C12N 9/00 (and C07K 14/00) are only used for the product (the inventive non-coding sequence of a gene) while C12N 15/00 is used for the use of this product (e.g. a promoter present in a vector for the production of other proteins). Non-coding sequences are only classified in C12N 9/00 (or C07K 14/00) if they are (part of) the invention. If the non-coding sequence is just an arbitrary choice from more available sequences it is not classified in C12N 9/00 (or C07K 14/00).
C-Sets classification:
In this group, C-Sets (#C12Na) are used. The detailed information about the C-Sets construction and the associated syntax rules are found in the "Special rules of classification" in C12N 15/10.
In this place, the following terms or expressions are used with the meaning indicated:
Operon | A DNA construct containing a cluster of genes under the control of a single regulatory signal or promoter |
This place covers:
Methods for generating mutants and their screening processes, without introducing genetic material into the organism (e.g by using chemical mutagens (nitrosoguanidine), specific culture conditions).
This place does not cover:
Methods for generating mutants by genetic engineering |
See also C12N 5/12 through C12N 5/166
This place covers:
This subclass will not be used.
This place covers:
Methods concerning the cloning, recombination, and generation of genetic material (DNA, RNA) by recombinant DNA technology; genetic engineering.
Attention is drawn to the following places, which may be of interest for search:
Host cells which are genetically engineered | |
Encoding nucleic acids, i.e. genes; Enzymes | |
Isolation of nucleic acids from host cells | C12N 15/1003, C12N 15/1006, C12N 15/101, C12N 15/1013, C12N 15/1017 |
DNA or RNA fragments; Modified forms thereof; Non-coding nucleic acids having biological activity, modulating the expression of genes (e.g. siRNA, miRNA,..; aptamers | |
Chemical preparation of DNA or RNA not used in recombinant technology | |
Encoding nucleic acids, i.e. genes; Polypeptides | |
Sequences of promoter, enhancer, intron, exon, IRES, IME are classified according to their corresponding gene sequence, e.g. polypeptide/enzyme | |
Preparation of non-structural polynucleotides from microorganisms or with enzymes | |
Nucleic acids used in diagnostics; Assays and products for analysing or detecting nucleic acids | |
Nucleic acid assays and products for analysing or detecting viruses or bacteriophages | |
Nucleic acids libraries |
Combination sets (C-Sets):
C-Sets statement : #C12Na
- In groups C12N 15/10 and its lower subgroups, DNA or RNA isolation/preparation processes are classified in the form of C-Sets.
- In these C-Sets, the base symbol, representing DNA or RNA isolation/preparation processes (or method), is taken from the groups C12N 15/10 - C12N 15/1096, whereas the subsequent symbol(s), representing the essential non-trivial technical feature(s) of the method, is (are) taken from the Indexing Codes under C12Q 2500/00 - C12Q 2565/634.
- Orthogonal symbols C12Q 2500/00 - C12Q 2565/634 are only used as subsequent symbols in C-Sets and should not be allocated as single symbols.
- In the C-set, only the essential technical features of the invention, which are non-trivial and differentiate them from the prior art, are to be represented using appropriate Indexing Codes C12Q 2500/00 - C12Q 2565/634.
- All indexing codes from groups C12Q 2500/00 - C12Q 2565/634 are to be used in the context literally expressed in the phrase ascribed to the code, i.e. the use of an indexing code is neither restricted by its hierarchical position in a group nor by the definition of the group in which the code is found.
C-Sets syntax rules:
- Each C-Set shall contain at least two symbols.
- Duplicate symbols are not allowed in these C-Sets. Multiple subsequent symbols from C12Q 2500/00 - C12Q 2565/634, if fit, can be used as C-Sets.
- The order of symbols in the C-Sets is relevant as it reflects the methods or process as a base symbol and the specific components of cell culture medium as a subsequent symbol, which are displayed in alphanumerical order.
C-Sets examples:
- #C12Na: A method of directed molecular evolution (C12N 15/1058) comprising incorporating random nucleotide sequences (C12Q 2525/179) by primer extension (C12Q 2533/101) of oligonucleotides comprising a modified backbone (C12Q 2525/113) is classified as (C12N 15/1058, C12Q 2525/113, C12Q 2525/179, C12Q 2533/101).
- #C12Na: A method of isolating genomic DNA by ion exchange (C12N 15/101) for the purpose of massive parallel sequencing (C12Q 2535/122) using a heat treatment (C12Q 2527/101) is classified as (C12N 15/101, C12Q 2527/101, C12Q 2535/122).
In patent documents, the following abbreviations are often used:
IRES | Internal Ribosome Entry Site |
IME | Intron Mediated Enhancement of gene Expression |
This place covers:
Methods for the isolation of nucleic acids e.g. DNA, RNA, plasmids, vectors, genomic DNA, genomic RNA) from microorganisms, viruses and plant origin.
This place does not cover:
Host cells which are genetically engineered | |
Chemical, physical, or physico-chemical processes in general; Their relevant apparatus | |
Solid sorbent composition or filter aid compositions per se, suitable for the isolation of nucleic acids | |
Ion exchange material per se, suitable for the isolation of nucleic acids | |
Any apparatuses per se, suitable for use in a process of extracting or isolating nucleic acids from microorganisms, viral or plant origin | |
Apparatuses for enzymology or microbiology per se, suitable for the extraction, isolation and preparation of nucleic acids |
This place covers:
Methods for the isolation of nucleic acids using a solid phase, e.g. non-magnetic beads.
This place does not cover:
Solid support carrier per se, suitable for carrying out the methods |
This place covers:
Methods for the isolation of nucleic acids by chromatography, binding the nucleic acid onto a column, washing and eluting the nucleic acid.
This place does not cover:
Chromatographic material per se, suitable for carrying out the methods |
This place covers:
Methods for the isolation of nucleic acids by chromatography, binding the nucleic acid to para-, ferro- or dia-magnetic beads, e.g. Dynabeads;
This place covers:
Methods for the isolation of nucleic acids by chromatography using filter, frits, membranes.
This place does not cover:
Filter material, frits and membranes per se, suitable for carrying out the methods |
This place covers:
General in vitro method of mutagenesis of nucleic acid by recombinant DNA technology, inserting/deleting/replacing nucleotides e.g. using oligonucleotides and site directed mutagenesis, error prone PCR, splicing by overlap extension.
This place does not cover:
DNA shuffling | |
Oligonucleotide assembly | |
Signature-tagged-mutagenesis (STM) |
See corresponding header for the C12N 15/10 group.
In patent documents, the following abbreviations are often used:
SOE | Splicing by Overlap Extension |
PCR | Polymerase Chain Reaction |
This place covers:
In vivo method of mutagenesis of nucleic acid by recombinant DNA technology, e.g. inserting/deleting/replacing nucleotides by introducing genetic material into the host cell, or disrupting the mismatch repair mechanism of the cell.
This place does not cover:
In vitro mutagenesis technique |
See corresponding header for the C12N 15/10 group.
This place covers:
- Mutagenesis by combined DNA shuffling;
- DNA sequence evolution by sexual PCR;
- Degenerate oligonucleotide gene shuffling and random drift mutagenesis.
This place does not cover:
In vitro mutagenesis by DNA fragment assembly |
See corresponding header for the C12N 15/10 group.
In patent documents, the following abbreviations are often used:
RSR | Recursive Sequence Recombination |
StEP | Staggered Extension Process |
PRP | Random-Priming in vitro Recombination |
ITCHY | Incremental Truncation for the Creation of Hybrid enzymes |
DOGS | Degenerate Oligonucleotide Gene Shuffling |
RNDM | Random Drift Mutagenesis |
This place covers:
Preparation of mutated genes by polynucleotide fragment assembly, e.g. assembly of genes using single-stranded oligonucleotides.
This place does not cover:
DNA shuffling |
See corresponding header for the C12N 15/10 group.
This place covers:
General method for screening libraries and isolating an individual clone in which the preparation of the library is not an essential technical feature.
Attention is drawn to the following places, which may be of interest for search:
Preparation of the library | |
Libraries per se | |
ICT specially adapted for in silico combinatorial libraries of nucleic acids, proteins or peptides | |
In silico combinatorial chemistry |
See corresponding header for the C12N 15/10 group.
Classification in C12N 15/1034 - C12N 15/1093 takes precedence over classification in C40B. That is to say C40B is of secondary importance and documents must always be classified in C12N 15/1034 and subgroups. A classification symbol from C40B may be given in addition. (N.B. C40B is never used for search in this field).
This place covers:
Preparation or screening of peptide libraries displayed by microorganism, cellular peptide display, e.g. phage display, E. coli display, yeast display, eukaryotic cell display.
Attention is drawn to the following places, which may be of interest for search:
Libraries per se contained in or displayed by microorganism |
See corresponding header for the C12N 15/1034 group.
This place covers:
Method for selecting high-affinity polypeptide ligands that specifically bind target molecules by ribosome/polysome display.
This place does not cover:
mRNA display | |
DNA display |
See corresponding header for the C12N 15/1034 group.
In patent documents, the following abbreviations are often used:
SPERT | Systematic Polypeptide Evolution by Reverse Translation |
ARM | Antibody-Ribosome-mRNA |
This place covers:
Preparation or screening a library of polypeptides comprising a scaffold-based molecule comprising loop domains, e.g. constructing or screening a library of a scaffold-based proteins which are derived from a stability enhanced consensus sequence of a fibronectin type III (FN3) domain incorporating randomized codons in order to produce polypeptide variants.
This place does not cover:
Library of scaffold based polypeptides |
See corresponding header for the C12N 15/1034 group.
This place covers:
- Method of aptamer selection, screening of DNA/RNA aptamers by systematic evolution of ligands by exponential amplification (SELEX);
- genomic SELEX;
- whole cell SELEX.
This place does not cover:
Aptamers per se | |
Peptide aptamers, i.e. build from aminoacids rather than nucleotidesamino acids rather than nucleotides | |
Library of aptamers |
See corresponding header for the C12N 15/1034 group.
In this place, the following terms or expressions are used with the meaning indicated:
SELEX | Systematic Evolution of Ligands by EXponential amplification |
Aptamers | Nucleic acid ligands |
This place covers:
Method of creating or screening a library of gene sequences by gene-trap cloning, isolation of gene sequences by gene-trap cloning, e.g. exons, introns, promoters, enhancer, signal sequences, IRES-, IME-sequences.
This place does not cover:
Preparation and screening expression libraries using reporter assays | |
Trap vectors per se |
See corresponding header for the C12N 15/10 and C12N 15/1034 groups.
In this place, the following terms or expressions are used with the meaning indicated:
IRES | Internal Ribosome Entry Site |
IME | sequence responsible for Intron Mediated Enhancement of gene expression |
This place covers:
- Preparation or screening libraries and isolating an individual clone which is involved in protein x protein interaction which involves gene expression, expression of a reporter gene e.g. in vivo library-versus-library selection of optimized protein-protein interactions, phage-based systems to select multiple protein-protein interactions simultaneously from combinatorial libraries,
- Yeast three-hybrid system,
- Three hybrid based screening assays using mammalian cells.
This place does not cover:
Preparation or screening expression libraries, e.g. reporter assays |
Attention is drawn to the following places, which may be of interest for search:
Methods of identifying protein-protein interactions in protein mixtures |
See corresponding header for the C12N 15/1034 group.
This place covers:
Method for the preparation, screening and isolation of gene sequences by a methods of directed molecular evolution using the modified polynucleotide libraries, e.g. cellular transformation, directed evolution, and screening methods for creating novel transgenic organisms having desirable properties, method of screening gene libraries derived from a mixed population of organisms for a bioactivity of biomolecule of interest.
See corresponding header for the C12N 15/1034 group.
This place covers:
Linking phenotype (polypeptide) covalently to mRNA (genotype) and screening libraries of such polypeptide-mRNA-display molecules for activity to identify single library member (polynucleotide sequence) that bind with a target molecule.
This place does not cover:
Polypeptide non-covalently bound to mRNA |
Attention is drawn to the following places, which may be of interest for search:
Cellular display | |
Ribosome/Polysome display | |
DNA display |
See corresponding header for the C12N 15/1034 group.
This place covers:
- Method of preparation or screening of tagged libraries;
- Method of tracking, identifying, and/or sorting classes or subpopulations of molecules by the use of (oligonucleotide) tags;
- Method whereby a molecular tag is put on a gene, transcript;
- Tag-creating DNA library, tagged microorganism by signature-tagged-mutagenesis (STM), e.g. gene identification signature (GIS) analysis, indexed library of cells, high throughput method for identification of sequence tags, tagged epitope protein transposable element, identification of transposon insertions within a transcribed portion of a gene of interest.
Attention is drawn to the following places, which may be of interest for search:
Library of templated molecules covalently or non-covalently linked to the encoded nuclei acid template, library of templated molecules linked to identifier oligonucleotides which have participated in the synthesis of said templated molecules. |
See corresponding header for the C12N 15/1034 group.
In this place, the following terms or expressions are used with the meaning indicated:
RBS-assay | Replica Barcode Selection assay |
GIS | Gene Identification Signature |
STM | Signature-Tagged-Mutagenesis |
This place covers:
- Methods and compositions for performing ordered multi-step synthesis by nucleic acid template mediated chemistry;
- Methods of synthesizing libraries of molecules comprising a functional moiety which is operatively linked to an encoding oligonucleotide;
- Library of templated molecules covalently or non-covalently linked to the encoded nuclei acid template;
- Library of templated molecules linked to identifier oligonucleotides which have participated in the synthesis of said templated molecules.
Attention is drawn to the following places, which may be of interest for search:
Libraries of templated molecules per se linked to their encoded nucleic acid templates |
See corresponding header for the C12N 15/1034 group.
This place covers:
- Preparation or screening at least two different libraries for obtaining or amplifying a polynucleotide (a tester-specific polynucleotide), in which an amount existing in a sample (tester) is larger than the amount existing in another sample (driver);
- Method to identify differentially expressed nucleotide sequences; subtractive hybridization process of enrichment of specific sequences from nucleic acid directory corresponding to a test directory; selective tagging of nucleic acids.
See corresponding header for the C12N 15/1034 group.
This place covers:
In vitro method of preparation or screening of libraries in cell-free compartmentalization systems (IVC), e.g. screening libraries prepared in emulsions (water in oil), droplets in contact with oil; directed evolution of polypeptide/enzyme (peptide display library) by in vitro compartmentalization;
covalent or non-covalent DNA-display.
This place does not cover:
Cellular display | |
Ribosome/Polysome display | |
mRNA display | |
Library of templated molecules covalently or non-covalently linked to the encoded nuclei acid template, library of templated molecules linked to identifier oligonucleotides which have participated in the synthesis of said templated molecules | |
Libraries per se |
Attention is drawn to the following places, which may be of interest for search:
Library of templated molecules covalently or non-covalently linked to the encoded nuclei acid template |
See corresponding header for the C12N 15/1034 group.
In this place, the following terms or expressions are used with the meaning indicated:
DNA display | Polypeptide is covalently or non-covalently bound to DNA |
IVC | In vitro cell-free compartmentalization systems |
This place covers:
Method of identifying nuclei acids that contributes to cell phenotype or phenotypic traits, e.g. method for the identification of genes that are essential for the maintenance of specific cell phenotypes.
See corresponding header for the C12N 15/1034 group.
This place covers:
Method of identifying nuclei acids by preparation or screening a genome library in which at least one nucleic acid was stable integrated e.g. using HR, site-specific recombination, transposons, viral vectors, into the genome of the host cell, e.g. using methods for site-specifically integrating at least one first nucleic acid into a genome of at least one cell.
See corresponding header for the C12N 15/1034 group.
In this place, the following terms or expressions are used with the meaning indicated:
HR | Homologous Recombination |
This place covers:
Method of identifying nuclei acids by preparation or screening a library of nucleic acids using reporter assays, wherein the reporter confers a selectable phenotype on cells, e.g. lacZ, GFP, YFP, Luc.
This place does not cover:
Gene-trapping | |
Libraries per se |
See corresponding header for the C12N 15/1034 group.
In this place, the following terms or expressions are used with the meaning indicated:
lacZ | Beta-galactosidase |
GFP | Green Fluorescent Protein |
YFP | Yellow Fluorescent Protein |
Luc | Luciferase |
This place covers:
Method of identifying nuclei acids by preparation or screening a library of nucleic acids in which at least one step comprises the use of a computer algorithm or an in silico step e.g. to align nucleotide sequences, in silico recombination techniques by designing oligonucleotides for regulated recombination.
This place does not cover:
Libraries per se |
Attention is drawn to the following places, which may be of interest for search:
ICT specially adapted for in silico combinatorial libraries of nucleic acids, proteins or peptides | |
In silico combinatorial chemistry |
See corresponding header for the C12N 15/1034 group.
This place covers:
Method of identifying nuclei acids by preparation or screening a library of nucleic acids/host cells not provided for in other subgroups.
See corresponding header for the C12N 15/1034 group.
This place covers:
Method for preparing cDNA by reverse translation of RNA, e.g. preparation and cloning DNA into a vector in which the reverse transcription is an essential feature of the process.
See corresponding header for the C12N 15/10 group.
This place covers:
Natural or synthetic nucleic acids used in biotechnology and genetic engineering.
Subgroups C12N 15/11 - C12N 15/117 also cover the use of non-coding nucleic acids as active ingredients medicinal preparations
This place does not cover:
DNA or RNA not used in recombinant technology. | |
Nucleotides and nucleosides per se and modified forms thereof. | |
Encoding nucleic acids, i.e. genes, with the exception of operons. |
Attention is drawn to the following places, which may be of interest for search:
Methods for isolating, preparing or purifying nucleic acids | |
Pharmaceutical compositions comprising nucleic acids | |
Use of medicinal preparations containing genetic material which is inserted into cells of the living body to treat genetic diseases, e.g. gene therapy | |
Chemical preparation of DNA or RNA not used in recombinant technology | |
Encoding nucleic acids, i.e. genes | |
Preparation of non-structural polynucleotides from microorganisms or with enzymes | |
Nucleic acids use in diagnostics; probes; primers | |
Nucleic acid libraries |
No distinction is made between 'invention' and 'additional' information. All technical features belonging to the 'invention' and all those exemplified in the document are classified. The allocation of additional information symbols, where possible, is mandatory.
Classification is made in all appropriate places, unless otherwise specified.
In this place, the following terms or expressions are used with the meaning indicated:
DNA | DeoxyriboNucleic Acid |
RNA | RiboNucleic Acid |
This place covers:
Methods and processes of general interest for one or more class(es) of non-coding nucleic acids.
This place does not cover:
The products per se |
Attention is drawn to the following places, which may be of interest for search:
Methods for isolating, preparing or purifying nucleic acids | |
Pharmaceutical compositions comprising nucleic acids | |
Use of medicinal preparations containing genetic material which is inserted into cells of the living body to treat genetic diseases, e.g. gene therapy | |
Chemical preparation of DNA or RNA not used in recombinant technology | |
Encoding nucleic acids, i.e. genes | |
Preparation of non-structural polynucleotides from microorganisms or with enzymes | |
Nucleic acids use in diagnostics; probes; primers | |
Nucleic acid libraries |
The methods or processes should always be further defined by the relevant codes of the C12N 2320/00 indexing scheme.
Methods or processes which are obviously state of the art are not classified
This place covers:
Nucleic acids having a direct effect on the expression of a gene or the transcription of its messenger such as Antisense nucleic acids, Catalytic nucleic acids, e.g. ribozymes, Triplex-forming oligonucleotides, Decoys, Nucleic acids used in gene silencing or RNA interference, or MicroRNAs.
This place does not cover:
Regulatory sequences being part of genes, e.g. promoters or terminators | |
Their use in plants | |
Encoding nucleic acids, i.e. genes | |
Probes and primers |
Attention is drawn to the following places, which may be of interest for search:
Methods for isolating, preparing or purifying nucleic acids | |
Pharmaceutical compositions comprising nucleic acids | |
Use of medicinal preparations containing genetic material which is inserted into cells of the living body to treat genetic diseases, e.g. gene therapy | |
Chemical preparation of DNA or RNA not used in recombinant technology | |
Encoding nucleic acids, i.e. genes | |
Preparation of non-structural polynucleotides from microorganisms or with enzymes | |
Nucleic acids use in diagnostics; probes; primers | |
Nucleic acid libraries |
In addition to the Rules already outlined for C12N 15/11, use of the C12N 2310/00 - C12N 2330/00 indexing schemes is made.
In this group and its subgroups classification is made according to the origin and nature of the target and should follow the categorisation made in C07K 14/00. Additional help can be found in the MeSH database of the NCBI
(www.ncbi.nlm.nih.gov/mesh ).
In this place, the following terms or expressions are used with the meaning indicated:
Antisense nucleic acid | DNA or RNA composed of the complementary sequence to the target DNA/RNA |
Ribozyme | Nucleic acid with a catalytic activity |
MicroRNA | Small double-stranded RNA (21-25 nucleotides) generated from a precursor transcript by cleavage with Dicer |
Decoy oligonucleotide | A (double-stranded) oligonucleotide comprising a binding-site for a protein |
Gene silencing | Interruption or suppression of the expression of a gene at transcriptional or translational levels. |
In patent documents, the following abbreviations are often used:
miRNA | MicroRNA |
TFO, triple helix | Triplex-forming oligonucleotide |
siRNA | Small interfering RNA |
shRNA | short hairpin RNA |
RNA interference | RNAi, gene interference, gene silencing, RNA silencing, cosuppression, co-suppression, post-transcriptional gene silencing, PTGS |
This place covers:
Non-coding nucleic acids directed against targets directly involved in the oncogenic processes, e.g. when their expression or activity is de-regulated.
In this place, the following terms or expressions are used with the meaning indicated:
Oncogenes | Genes whose gain-of-function alterations lead to neoplastic cell transformation |
Tumour suppressor genes | Genes that inhibit formation and/or development of the tumorigenic phenotype |
This place covers:
Non-coding nucleic acids directed to nucleic acids encoding proteins with an enzymatic activity.
This place does not cover:
Viral enzymes | |
Receptors having also an enzymatic domain |
The enzyme(s) targeted are further specified by using the appropriate symbol(s) from the C12Y scheme.
This place covers:
- Nucleic acids targeting receptors, i.e. membrane-embedded proteins transmitting a signal after binding of their ligand(s), including nuclear receptors.
- Nucleic acids targeting any protein normally present on the surface of a cell, e.g. ion channels.
This place does not cover:
Nucleic acids against binding proteins other than receptors |
This place covers:
Nucleic acids binding a target molecule specifically and with high affinity without hybridising therewith.
This place does not cover:
Peptide aptamers, i.e. built from aminoacids rather than nucleotides. |
Attention is drawn to the following places, which may be of interest for search:
Nucleic acid libraries |
- The corresponding Indexing Code should always be given in addition to the class.
- Known aptamers linked to another entity as regulatory domain should be classified in the group corresponding to said entity and the Indexing Code for the aptamer added.
In patent documents, the following abbreviations are often used:
Aptamer | Nucleic acid ligand, NAL |
This place covers:
Non-coding nucleic acids having a direct impact on the immune system.
Attention is drawn to the following places, which may be of interest for search:
Adjuvants containing nucleic acids |
The corresponding Indexing Code should always be given in addition to the class.
This place covers:
Operons: DNA constructs containing a cluster of genes under the control of a single regulatory signal or promoter.
Groups of genes for enzymes providing an organism with the ability to synthesize a specific compound or compounds (e.g. synthetic (partial) pathways)
This place does not cover:
Genes coding for single enzymes |
This place covers:
Methods to produce a fusion protein by recombinant DNA technology
This place does not cover:
Fusion proteins per se and the nucleic acids encoding them. |
This group covers only documents in which emphasis is given on the method for the preparation of fusion proteins.
The products of the method are also classified in the groups for the individual proteins being part of the fusion protein.
The documents have further to be given an Indexing Code for fusion proteins: C07K 2319/00.
This place covers:
Methods for producing, by recombinant DNA technology, a fusion protein in which one of the fused polypeptides consists of a signal sequence with or without (part of) its mature protein.
This place does not cover:
Fusion proteins per se |
This group covers only documents in which emphasis is given on the method for the preparation of fusion proteins containing a signal sequence.
The products of the method are also classified in the groups for the individual proteins being part of the fusion protein. This includes classification into the group for the polypeptide from which the signal sequence has been derived
The document has further to be given an Indexing Code for fusion proteins: C07K 2319/00.
In this place, the following terms or expressions are used with the meaning indicated:
Fusion protein | A polypeptide consisting of (parts of) two or more different proteins covalently linked to each other by a peptide bond |
Signal sequence | A 3-60 amino acid long peptide that directs the transport of the protein that is attached to it |
This place covers:
General method for regulating (enhancing, inhibiting) the gene expression by modifying the operator, enhancer or promoter dependent transcription of the messenger RNA.
This place does not cover:
Ribosome mediated translational regulation of gene expression |
See corresponding header for the C12N 15/00 group.
This place covers:
General method for regulating (enhancing, inhibiting) the gene expression by modifying/regulating the repressor or inducer mediated transcription of the messenger RNA.
This place does not cover:
Ribosome mediated translational regulation of gene expression |
See corresponding header for the C12N 15/00 group.
This place covers:
General methods for generating vectors for use in recombinant technology i.e. cloning methods for preparing a general vector. A general vector is to be understood as being one which is independent of the origin of replication. Methods for specifically preparing e.g. a plant, viral or mammalian vector, wherein the method of preparation is restricted to and applicable only in e.g. a plant, virus or mammal are classified in their corresponding vector group.
Attention is drawn to the following places, which may be of interest for search:
Bacterial vectors | C12N 15/70, C12N 15/71, C12N 15/72, C12N 15/73, C12N 15/74, C12N 15/75, C12N 15/76, C12N 15/77, C12N 15/78 |
Eukaryotic vectors | |
Fungal vectors | |
Plant vectors | |
Animal vectors | |
Viral vectors for animal cells |
Combination sets (C-Sets):
C-Sets statement : #C12Nb
- In groups C12N 15/64 - C12N 15/66, methods for preparing vectors are classified in the form of C-Sets.
- In these C-Sets, the base symbol, representing methods for preparing vectors, is taken from the groups C12N 15/64 - C12N 15/66, whereas the subsequent symbols, representing the feature(s) of the methods, is/are taken from the Indexing Codes under C12Q 2500/00 - C12Q 2565/634.
- Orthogonal symbols C12Q 2500/00 - C12Q 2565/634 are only used as subsequent symbols in C-Sets and should not be allocated as single symbols.
- In the C-set, only the essential technical features of the invention, which differentiate it from the prior art, are to be represented.
- All indexing codes from groups C12Q 2500/00 - C12Q 2565/634 are to be used in the context literally expressed in the phrase ascribed to the code, i.e. the use of an indexing code is neither restricted by its hierarchical position in a group nor by the definition of the group in which the code is found.
C-Sets syntax rules:
- Each C-Set shall contain at least two symbols.
- Duplicate symbols are not allowed in these C-Sets.
- The order of symbols in the C-Sets is relevant as it reflects the methods or process as a base symbol and as a subsequent symbol, which are displayed in alphanumerical order.
C-Sets examples:
- #C12Nb: A method for inserting a gene of interest into a vector (C12N 15/66) using a type IIS restriction enzyme (C12Q 2521/313), and a ligase (C12Q 2521/501) to attach an adaptor (C12Q 2525/191) at each extremity of the gene of interest is classified as (C12N 15/66, C12Q 2521/313, C12Q 2521/501, C12Q 2525/191).
- #C12Nb: A method for preparing a vector (C12N 15/64) comprising amplifying a template using strand-displacement rolling circle (C12Q 2531/125) is classified as (C12N 15/64, C12Q 2531/125).
This place covers:
General methods for preparing the vector, for introducing it into the cell or for selecting the vector-containing host using polypeptides as marker molecules, wherein the polypeptides do not comprise enzymatic activity.
This place does not cover:
Enzymes used as marker molecules |
See corresponding header for the C12N 15/00 group.
See corresponding header for the C12N 15/10 group.
The methods or processes should always be classified in combination-sets which consist of the appropriate CPC group together with the Indexing Codes under C12N 2500/00.
This place covers:
General method for preparing a recombinant vector using cleavage and ligation.
See corresponding header for the C12N 15/00 group.
See corresponding header for the C12N 15/10 group.
The methods or processes should always be classified in combination-sets which consist of the appropriate CPC group together with the Indexing Codes under C12N 2500/00.
In this place, the following terms or expressions are used with the meaning indicated:
Non-functional linkers | DNA sequences which are used to link DNA sequences and which have no known function of structural gene or regulating function. |
This place covers:
General method for ribosome mediated regulating (i.e. enhancing, inhibiting) the gene expression by modifying the ribosomal mediated translation of the messenger RNA.
This place does not cover:
Transcriptional regulation of gene expression |
This place covers:
General method of enhancing the expression a nucleic acid sequence, by stabilizing the vector in the host cell, e.g. preserving DNA in a stable form over time, temperature, culture conditions.
This place covers:
General method of enhancing the expression a nucleic acid sequence, by increasing the copy number of the vector in the host cell, e.g. conditions that results in an increase in plasmid copy number in comparison to a control plasmid, e.g. mutation in the copy number control region.
This place covers:
All application dealing with vectors especially adapted for E.coli, i.e. comprising at least one origin of replication working in Escherichia coli.
Shuttle vector is classified according to the vector/host system in which the vector is able to replicate.
Vectors comprising a chimeric/hybrid origin of replication are classified according to the vectors/host system in which the vector is able to replicate.
In this place, the following terms or expressions are used with the meaning indicated:
Shuttle vector | Vector comprising at least two different origins of replication |
This place covers:
All application dealing with vectors especially adapted for E.coli, i.e. comprising at least one origin of replication working in Escherichia coli, and comprising regulatory sequences from the trp-operon.
See corresponding header for C12N 15/70.
In this place, the following terms or expressions are used with the meaning indicated:
Trp-operon | Tryprophan-operon |
Operon | Set of neighbouring prokaryotic genes whose transcription is simultaneously controlled |
This place covers:
All application dealing with vectors especially adapted for E.coli, i.e. comprising at least one origin of replication working in Escherichia coli, and comprising regulatory sequences from the lac-operon.
See corresponding header for C12N 15/70.
In this place, the following terms or expressions are used with the meaning indicated:
Lac-operon | Lactose-operon |
This place covers:
All application dealing with vectors especially adapted for E.coli, i.e. comprising at least one origin of replication working in Escherichia coli, and comprising regulatory sequences from the phage (lambda) regulatory sequences.
See corresponding header for C12N 15/70.
This place covers:
All application dealing with vectors especially adapted for prokaryotic hosts other than E.coli, i.e. comprising at last one origin of replication working in prokaryotic hosts other than E. coli, e.g. Lactobacillus, Micromonospora.
See corresponding header for C12N 15/70.
This place covers:
All application dealing with vectors especially adapted for prokaryotic hosts other than E.coli, i.e. comprising at least one origin of replication working in prokaryotic hosts other than E. coli, e.g. for Agrobacterium, Rhizobium, Bradyrhizobium.
See corresponding header for C12N 15/70.
This place covers:
All application dealing with vectors especially adapted for prokaryotic hosts other than E.coli, i.e. comprising at least one origin of replication working in prokaryotic hosts other than E. coli, e.g.: for lactic acid bacteria (Streptococcus; Lactococcus; Lactobacillus; Pediococcus; Enterococcus; Leuconostoc; Propionibacterium; Bifidobacterium; Sporolactobacillus).
See corresponding header for C12N 15/70.
This place covers:
All application dealing with vectors especially adapted for prokaryotic hosts other than E.coli, i.e. comprising at least one origin of replication working in prokaryotic hosts other than E. coli, e. g. for Bacillus.
See corresponding header fro C12N 15/70.
This place covers:
All application dealing with vectors especially adapted for prokaryotic hosts other than E.coli, i.e. comprising at least one origin of replication working in prokaryotic hosts other than E. coli, e. g. for Actinomyces; for Streptomyces.
See corresponding header for C12N 15/70.
This place covers:
All application dealing with vectors especially adapted for prokaryotic hosts other than E.coli, i.e. comprising at least one origin of replication working in prokaryotic hosts other than E. coli, e. g. Corynebacterium, for Brevibacterium.
See corresponding header for C12N 15/70.
This place covers:
All application dealing with vectors especially adapted for prokaryotic hosts other than E.coli, i.e. comprising at least one origin of replication working in prokaryotic hosts other than E. coli, e. g. Pseudomonas.
See corresponding header for C12N 15/70.
This place covers:
All application dealing with vectors especially adapted for eukaryotic hosts, i.e. comprising at least one origin of replication working in eukaryotic hosts, and which can not be classified in the subgroups below C12N 15/80 – C12N 15/86.
See corresponding header for C12N 15/70.
See corresponding header for C12N 15/64.
This place covers:
All application dealing with vectors especially adapted for eukaryotic hosts, i.e. comprising at least origin of replication working in eukaryotic hosts, e.g. fungi.
See corresponding header for C12N 15/70.
See corresponding header for C12N 15/64.
This place covers:
All application dealing with vectors especially adapted for eukaryotic hosts, i.e. comprising at least one origin of replication working in eukaryotic hosts, e.g. yeast.
See corresponding header for C12N 15/70.
See corresponding header for C12N 15/64.
This place covers:
All application dealing with vectors especially adapted for eukaryotic hosts, i.e. comprising at least one origin of replication working in eukaryotic hosts, e.g. other than Saccharomyces.
See corresponding header for C12N 15/70.
See corresponding header for C12N 15/64.
This place covers:
- New transgenic plants, methods, vectors, constructs, promoters, etc. for transforming, expressing;
- Modifying genotypes/phenotypes of plants by genetic engineering.
If a document relates to:
Plant breeding (tissue culture) method and new non-transgenic plants (varieties): use A01H 1/00 through A01H 4/00 for the method if appropriate; use A01H 5/00 through A01H 17/00 for the new plants (varieties)
- Purposive modification of plant phenotype/genotype (e.g. method for altering starch composition), creation of transgenic plants and transgenic plants themselves: use C12N 15/8241... subclass for the method if appropriate;use C12N 15/8241... subclass as appropriate for the plant if transgenic normally no A01H class (A01H class could be given if there is information regarding the transgenic plant as a variety)
- Transformation method or expression method (including constructs, promoters etc): use C12N 15/8201... or C12N 15/8216... for the methods as appropriate
Note that the C12N 15/8241... subclasses are not used unless there is significant matter defining the purposive modification of the plant genotype or phenotype normally no A01H class as above. (Note also A01H 1/00 through A01H 4/00 could be added. For example if transformation method included information relating to tissue culture).
A01H 5/00 through A01H 17/00 classes are only used in ECLA for new mainly non-transgenic plants (usually varieties).
Includes algal transformation
This place covers:
Means for DNA/chromosomal rearrangements
This place covers:
Mitochondrial transformation
In this place, the following terms or expressions are used with the meaning indicated:
Output trait influences the output products of plants or their parts
This place covers:
Modulating protein content
In this place, the following terms or expressions are used with the meaning indicated:
Input trait influences the input required for growth and development of the plant or its parts
This place covers:
Vectors are nucleic acid constructs capable of introducing genetic information into cells.This group covers vectors for animal cells (C12N 15/85), in particular for the production of transgenic animals (C12N 15/8509), and viral vectors (C12N 15/86).
Introduction of foreign genetic material using processes not otherwise provided for, e.g. co-transformation, cloning and microinjection are covered by C12N 15/87-C12N 15/907.
C12N 15/8509, C12N 15/873 and C12N 15/877 relate to processes for producing animals, which per se are classified in the range A01K 67/027-A01K 67/60, in combination with codes in the A01K 2207/00-A01K 2267/00 ranges.
This place does not cover:
Vectors specialised for bacterial cells | |
Vectors specialised for yeast cells | |
Vectors specialised for plant cells |
- For all vectors listed below, the relevant uses and functional elements should be indicated using codes in the C12N 2800/00-C12N 2840/00 ranges unless indicated otherwise.
- C12N 15/85 is to be given when a vector is particularly suitable for introduction of genetic material into animal cells.
- C12N 15/8509 is to be combined with codes in the A01K 2267/00 series to indicate the particular purpose of the produced animal model. A01K 67/027-A01K 67/60 is to be given if the animal has actually been generated. In these cases, (a) code(s) in the A01K 2217/00 range are to be used to further define the vector and animal. With respect to the use of codes in the C12N 2830/00 range in combination with C12N 15/8509 classes, A01K 2217/203 takes precedence over C12N 2830/001 and C12N 2830/007, and A01K 2217/206 takes precedence over C12N 2830/008. The C12N 2830/001-C12N 2830/008 codes are only to be given when the invention actually resides in these elements. C12N27xx/xxx43 (where x is an integer from 0 to 9) codes are to be given to combine viral taxonomy with vector use in generating genetically modified animals.
- The subdivision of viral vectors corresponding to IPC C12N 15/86 and subclasses is no longer used. It is mandatory to give symbols from the C12N 2710/00-C12N 2795/00 series combining the viral taxonomy with the appropriate ending 43, 44 or 45 to indicate subject-matter relating to viral vectors. When the viral vector is an essential part of the invention, C12N 15/86 needs to be co-allocated.
- C12N 15/87 and C12N 15/88 relate to introduction of foreign genetic material by processes not otherwise provided for, whereby the vectors of C12N 15/88 are microencapsulated. If such subject-matter is for therapeutic use, gene therapy (A61K 48/00) and relevant non-active ingredients need to be considered (A61K 47/00). For microencapsulation, the liposome composition may be of relevance (A61K 9/127).
- Processes for the production of new or cloned embryos are to be classified in C12N 15/873 and C12N 15/877. If new animals have been actually produced, also the relevant A01K 67/027-A01K 67/60 class and relevant codes in the A01K 2207/00-A01K 2267/00 ranges are to be given. For the manipulation of cells, C12N 5/00 groups should be considered.
- C12N 15/89 and subgroups relate to processes for introduction of foreign genetic material by microinjection, in particular using biolistic methods (C12N 15/895).
- Vectors for homologous recombination are classified in C12N 15/902, with specific subdivision for yeast (C12N 15/905) and mammalian cells (C12N 15/907). The last place rule applies.
- Introduction of genetic material specifically into plant cells should also be classified in C12N 15/82 and subgroups.
This place covers:
All indexing information needed to characterise the non-coding nucleic acids used in groups C12N 15/11 - C12N 15/117.
Attention is drawn to the following places, which may be of interest for search:
Specific uses or applications | |
Ways of production or obtention |
The orthogonal Indexing symbols in this group are only to be used with groups C12N 15/11 - C12N 15/117.
The orthogonal Indexing symbols are only given to information relevant to the invention or explicitly exemplified, i.e. wish-lists are not indexed.
C-Sets classification:
C-Sets statement: #C12Nc
- In groups C12N 2310/00 - C12N 2310/533, a combination of features characterising the structure or type of the nucleic acid can be classified in the form of C-Sets when appropriate.
- In these C-Sets, the base symbol, representing the structure or type of the nucleic acid, is taken from the groups C12N 2310/00 - C12N 2310/533, followed by a subsequent symbol representing a further characteristic of said nucleic acid is selected from the groups C12N 2310/00 - C12N 2330/51.
C-Sets syntax rules:
- Each C-Set shall contain at least two symbols.
- Duplicate symbols are not allowed in these C-Sets.
- The C-Sets is allocated as ADD.
- The order of symbols in these C-Sets is not relevant.
C-Sets Examples:
#C12Nc: a nucleic acid that is modified at the 2'-OR position of the sugar by a methoxy-group: is classified as (C12N 2310/321, C12N 2310/3521)
#C12Nc: a nucleic acid that is halogenated at the 2'-R-position of the sugar is classified as (C12N 2310/322, C12N 2310/3533)
#C12Nc: a methylated adenosine is classified as (C12N 2310/333, C12N 2310/3521)
#C12Nc: an interfering RNA with a stem-loop structure, e.g. a shRNA is classified as (C12N 2310/14, C12N 2310/531)
#C12Nc: an antisense nucleic acid with an LNA - gapmer design is classified as (C12N 2310/11, C12N 2310/3231, C12N 2310/341)
C-Sets searches:
C-Sets search queries may be made according to C-Sets classification rule #C12Nc described above.
This place covers:
Information about which class of non-coding nucleic acid is concerned.
Attention is drawn to the following places, which may be of interest for search:
The chemical structure of the nucleic acids | |
The physical structure of the nucleic acids |
Attention is drawn to the following places, which may be of interest for search:
Structure or type of nucleic acid, Aptamers | |
Structure or type of Immunomodulatory nucleic acids |
This place covers:
Chemical modification of the nucleic acids, i.e. all relevant deviations from the natural DNA or RNA forms.
Attention is drawn to the following places, which may be of interest for search:
Type of nucleic acids | |
The physical structure of the nucleic acids | |
Chemical modification of nucleotides and nucleosides |
This place covers:
Modifications of the phosphate group(s) forming the backbone of the nucleic acid, including the terminal group(s).
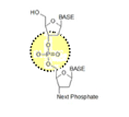
In this place, the following terms or expressions are used with the meaning indicated:
Phosphonate | O=P(O)2-R |
Phosphorodithioate | S=P(O)2-SR, S=P(SO)-OR |
Phosphorothioate | S=P(O)2-OR, O=P(O)2-SR |
Phosphoramidate | O=P(O)2-NR2, O=P(NO)-OR |
Phosphotriester | O=P(O)2-O-R |
Phosphonothioate | S= P(O)2-R |
This place covers:
Nucleic acids where the phosphate units of the backbone are (at least partially) replaced by a different chemical entity.
This place covers:
All modifications made to, or variations of, the (deoxy)ribose part of the nucleic acid:
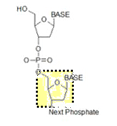
This place covers:
All modifications at the 2'-position of the sugar made via an oxygen-atom.
Attention is drawn to the following places, which may be of interest for search:
Modified sugar ring structures |
The nature of the modifying group is further defined by using a combination symbol (see the corresponding header of C12N 2310/00).
The combination symbol corresponding to the natural ribose (2'-OH), i.e. C12N 2310/321 combined with C12N 2310/3531 to form a combination set is only given when the RNA nature of the oligonucleotide is of particular relevance to the invention.
This place covers:
All modifications directly linked to the 2'-carbon of the sugar, without an intermediate oxygen.
Attention is drawn to the following places, which may be of interest for search:
Modified sugar ring structures |
The nature of the modifying group is further defined by using a combination symbol (see the corresponding header of C12N 2310/00).
The combination symbol corresponding to the natural deoxyribose (2'-H), i.e. C12N 2310/322 combined with C12N 2310/3531 to form a combination set is only given when the DNA nature of the oligonucleotide is of particular relevance to the invention.
This place covers:
Modifications of the classical (deoxy)ribose ring, including replacement by other sugars.
Attention is drawn to the following places, which may be of interest for search:
Modifications made at positions of the sugar not affecting the (deoxy)ribose nature |
The nature of the modifying group is further defined by using a combination symbol (see the corresponding header of C12N 2310/00).
This place covers:
Nucleic acids comprising a base other than adenine (A), uracil (U), thymidine (T), guanine (G) or cytosine (C). This includes modifications of said natural bases.
Attention is drawn to the following places, which may be of interest for search:
Nucleic acids where one or more base(s) is (are) missing |
The nature of the modifying group is further defined by using a combination symbol (see the corresponding header of C12N 2310/00).
This place covers:
Nucleic acids where the specific position of a modification within the nucleic acid is relevant.
Attention is drawn to the following places, which may be of interest for search:
Modifications of the backbone, sugars or bases per se |
This place covers:
Aspects concerning specific uses of the non-coding nucleic acids.
This place does not cover:
Structural aspects of the nucleic acids | |
Methods for producing or obtaining the nucleic acids |
Attention is drawn to the following places, which may be of interest for search:
2ND medical uses | |
Gene therapy |
The indexing symbols in this group are only to be used with groups C12N 15/11 - C12N 15/117.
The indexing symbols are only given to information relevant to the invention or explicitly exemplified, i.e. wish-lists are not indexed.
When appropriate, use of combination-symbols is made to further characterise the nucleic acids (see corresponding rule by C12N 2310/00).
This place covers:
The use of the non-coding nucleic acid for screening as well as the detection of the nucleic acid.
Attention is drawn to the following places, which may be of interest for search:
Use of nucleic acids in screening |
This place covers:
The use of a non-coding nucleic acid in the identification of an accessible site on the target nucleic acid. Hence, the Indexing Code covers also the screening for active non-coding nucleic acids.
Attention is drawn to the following places, which may be of interest for search:
The use in functional genomics | |
Assays involving nucleic acids |
This place covers:
All methods and processes directed to provide the non-coding nucleic acid with a new function.
Attention is drawn to the following places, which may be of interest for search:
SELEX |
This place covers:
Subject-matter where the method or therapeutic application is part of the invention.
Attention is drawn to the following places, which may be of interest for search:
Pharmaceutical compositions comprising the nucleic acids; second therapeutic applications | |
Gene therapy |
This place covers:
Methods and means directed to modify the natural activity of the nucleic acids.
This place does not cover:
The directed acquisition of a totally new activity |
This place covers:
The means for producing or obtaining the non-coding nucleic acids.
This place does not cover:
The structural aspects of the nucleic acids | |
Specific uses |
Attention is drawn to the following places, which may be of interest for search:
Methods for isolating or producing nucleic acids in general |
The indexing symbols in this group are only to be used with groups C12N 15/11 - C12N 15/117.
The indexing symbols are only given to information relevant to the invention or explicitly exemplified, i.e. wish-lists are not indexed.
When appropriate, use of combination-symbols is made to further characterise the nucleic acids (see corresponding rule by C12N 2310/00).
This place covers:
Non-coding nucleic acids naturally present in a cell or organism.
This Indexing Code can be given as a combination code, e.g. C12N 2310/111 combined with C12N 2330/10 to form a combination set indicates a naturally occurring antisense.
Attention is drawn to the following places, which may be of interest for search:
Vitamins of cell culture medium |
Attention is drawn to the following places, which may be of interest for search:
Insulin used in cell culture processes, e.g. differentiation |
Attention is drawn to the following places, which may be of interest for search:
Metals; Metal chelators | |
Light metals, Calcium; Ca chelators; Calcitonin | |
Transition metals, Iron; Fe chelators; Transferrin |
Attention is drawn to the following places, which may be of interest for search:
Modulators of cAMP or cGMP, e.g. non-hydrolysable analogues, phosphodiesterase inhibitors, cholera toxin | |
Anti-neoplasic drugs, anti-retroviral drugs, e.g. azacytidine, cyclophosphamide |
Attention is drawn to the following places, which may be of interest for search:
Conditioned medium |
Attention is drawn to the following places, which may be of interest for search:
Hormones derived from pro-opiomelanocortin, pro-enkephalin or pro-dynorphin |
Attention is drawn to the following places, which may be of interest for search:
Insulin-transferrin; Insulin-transferrin-selenium | |
Insulin-like growth factors [IGF] |
Attention is drawn to the following places, which may be of interest for search:
Modulators of histone acetylation |
Attention is drawn to the following places, which may be of interest for search:
Coating allowing for selective detachment of cells, e.g. thermoreactive coating |