| US 11,817,214 B1 | ||
| Machine learning model trained to determine a biochemical state and/or medical condition using DNA epigenetic data | ||
| Jon Sabes, Minneapolis, MN (US); Brian H. Chen, Minneapolis, MN (US); and Randal S. Olson, Minneapolis, MN (US) | ||
| Assigned to FOXO Labs Inc., Minneapolis, MN (US) | ||
| Filed by Life Epigenetics, Inc., Minneapolis, MN (US) | ||
| Filed on Sep. 23, 2019, as Appl. No. 16/579,818. | ||
| Int. Cl. G06K 9/00 (2018.01); G16H 50/20 (2018.01); G06N 20/00 (2019.01); G06Q 40/08 (2012.01); G16B 20/00 (2019.01); G16B 40/30 (2019.01); G16B 5/00 (2019.01); G16B 40/20 (2019.01) | ||
| CPC G16H 50/20 (2018.01) [G06N 20/00 (2019.01); G06Q 40/08 (2013.01); G16B 5/00 (2019.02); G16B 20/00 (2019.02); G16B 40/20 (2019.02); G16B 40/30 (2019.02)] | 20 Claims |
|
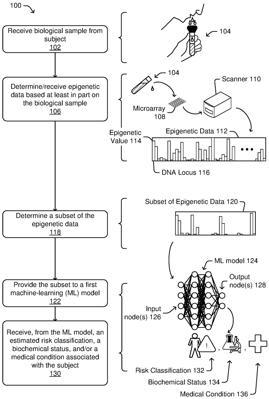
1. A method comprising:
training a machine-learning (ML) model to estimate a biochemical state or a medical condition associated with a first subject based at least in part on epigenetic data associated with a biological sample received from the first subject, wherein the training comprises:
receiving a plurality of training sets of epigenetic data associated with different subjects, the epigenetic data being associated with a plurality of DNA loci;
receiving a plurality of labels associated with the plurality of training sets, wherein a first label of the plurality of labels is associated with a second subject and comprises a classification or value associated with the biochemical state or the medical condition;
determining, based at least in part on a feature reduction algorithm, a subset of DNA loci from among the plurality of DNA loci; and
training the ML model based at least in part on:
providing, to one or more input nodes of the ML model, a portion of the plurality of training sets of epigenetic data corresponding to the subset of DNA loci;
determining, by one or more hidden layers based at least in part on providing the portion to the one or more input nodes, an intermediate output;
determining, by an output layer based at least in part on the intermediate output, an output; and
altering a parameter of the ML model based at least in part on a loss determined between the output of the ML model and at least one of the one or more label values, wherein the output is based at least in part on the first training epigenetic values.
|