| CPC G16C 20/10 (2019.02) [G06N 3/04 (2013.01); G06N 3/08 (2013.01)] | 20 Claims |
|
1. A computational method for simulating motion of elements within a multi-element system using a graph neural network (GNN), the computational method comprising:
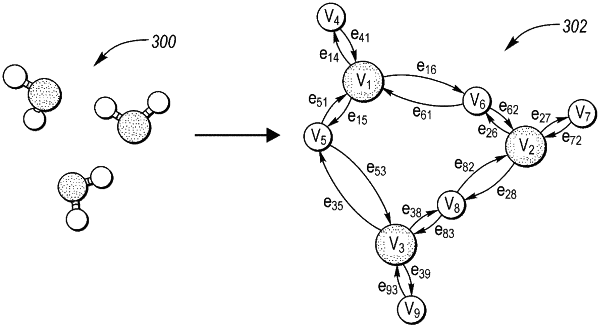
converting a molecular dynamics snapshot of the elements within the multi-element system into a directed graph comprised of nodes and edges, the nodes including first and second nodes and the edges including first and second edges;
initially embedding the nodes and the edges to obtain initially embedded nodes and edges;
updating the initially embedded nodes and edges by passing a first message from the first edge to the first node using a first message function and passing a second message from the first node to the first edge using a second message function to obtain updated embedded nodes and edges, the first message function uses the first message and the initially embedded nodes as inputs, the first message function concatenates the initially embedded nodes, with a hyperbolic tangent function and a sigmoid function applied to the result to obtain first and second outputs;
predicting a force vector for one or more elements within the multi-element system based on the updated embedded edges and a unit vector pointing from the first node to the second node or the second node to the first node; and
training the GNN to update weights and biases through backpropagation to extract one or more rotationally-invariant features from the molecular dynamics snapshot to predict atomic forces to reduce error between the predicted forces and the molecular dynamics calculated forces.
|