| CPC G16B 15/30 (2019.02) [G16B 40/00 (2019.02); G16B 45/00 (2019.02)] | 30 Claims |
|
1. A method for the in-silico design of an amino acid interface of a biologic for binding to a target, the method comprising:
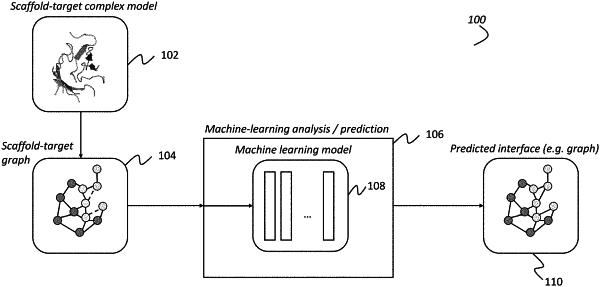
(a) receiving, by a processor of a computing device, an initial scaffold-target complex graph comprising a graph representation of at least a portion of a biologic complex comprising the target and a peptide backbone of the in-progress custom biologic, the initial scaffold-target complex graph comprising:
a target graph representing at least a portion of the target; and
a scaffold graph representing at least a portion of the peptide backbone of the in-progress custom biologic, the scaffold graph comprising a plurality of scaffold nodes, a subset of which are unknown interface nodes, wherein each of said unknown interface nodes:
(i) represents a particular amino acid interface site, along the peptide backbone of the in-progress custom biologic, that is located in proximity to one or more amino acids of the target, and
(ii) has a corresponding node feature vector comprising a side chain type component vector populated with one or more masking values, thereby representing an unknown, to-be determined, amino acid side chain;
(b) generating, by the processor, using a machine learning model, one or more likelihood graphs based on the initial scaffold-target complex graph, each of the one or more likelihood graphs comprising a plurality of nodes, a subset of which are classified interface nodes, each of which:
(i) corresponds to a particular unknown interface node of the scaffold graph and represents a same particular interface site along the peptide backbone of the in-progress custom biologic as the corresponding particular interface node, and
(ii) has a corresponding node feature vector comprising a side chain component vector populated with one or more likelihood values;
(c) using, by the processor, the one or more likelihood graphs to determine a predicted interface comprising, for each interface site, an identification of a particular amino acid side chain type; and,
(d) providing the predicted interface for use in designing the amino acid interface of the in-progress custom biologic and/or using the predicted interface to design the amino acid interface of the in-progress custom biologic.
|