| CPC G16H 10/60 (2018.01) [G16H 15/00 (2018.01); G16H 20/40 (2018.01); G16H 50/20 (2018.01); G16H 50/30 (2018.01); G16H 50/50 (2018.01); G16H 50/70 (2018.01)] | 86 Claims |
|
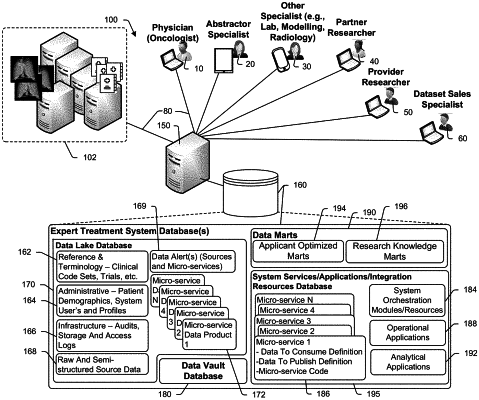
1. A method for conducting genomic sequencing, the method comprising the steps of:
storing a set of user application programs wherein each of the programs requires an application specific subset of data to perform application processes and generate user output;
storing a plurality of micro-service programs;
for each of a plurality of patients that have cancerous cells and that receive cancer treatment:
a. obtaining clinical records data in original forms where the clinical records data includes cancer state information, treatment types and treatment efficacy information;
b. storing the clinical records data in a semi-structured first database, the storing including identifying one or more of a document type or source of the clinical records, the storing further including extracting data from the clinical records using a machine learning algorithm (MLA), deep learning algorithm (DLA), or neural network (NN), wherein the MLA, DLA, or NN is selected from among a plurality of MLAs, DLAs, and/or NNs based upon the identified one or more of the document type or source of the clinical records;
c. for each patient, using a next generation genomic sequencer to generate DNA and whole-transcriptome RNA genomic sequencing data from the patient's cancerous cells and normal cells;
d. storing the sequencing data in the first database;
e. shaping at least a subset of the first database data to generate system structured data including clinical record data and sequencing data wherein the system structured data is optimized for searching;
f. storing the system structured data in a second database;
g. for each user application program:
i. selecting the application specific subset of data from the second database; and
ii. storing the application specific subset of data in a structure optimized for application program interfacing in a third database; and
wherein an orchestration manager is operatively connected to one or more micro-service programs to receive status messages from micro-service programs and initiate a respective micro-service program when prerequisites of that respective micro-service program are satisfied.
|