| CPC G06T 1/20 (2013.01) [G06F 18/21 (2023.01); G06F 18/2431 (2023.01); G06T 7/0012 (2013.01); G06T 7/11 (2017.01); G06T 11/00 (2013.01); G06V 10/44 (2022.01); G06V 10/764 (2022.01); G06V 10/82 (2022.01); G06T 2207/20081 (2013.01); G06T 2207/30024 (2013.01); G06T 2207/30096 (2013.01)] | 28 Claims |
|
1. A computer-implemented method of identifying biomarkers in a digital image of a hematoxylin and eosin (H&E) stained slide of target tissue, the method comprising:
receiving the digital image to an image-based biomarker prediction system having one or more processors;
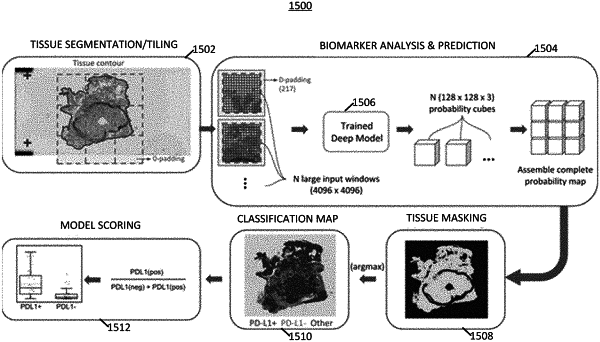
performing an image tiling process, using the one or more processors, on the digital image by separating the digital image into a plurality of tile images, where each of the plurality of tile images contains a different portion of the digital image;
applying, using the one or more processors, the plurality of tile images to a multiscale deep learning framework comprising one or more trained deep learning multiscale classifier models, each trained deep learning multiscale classifier models being trained to classify a different tissue classification for each tile image and determining a tissue classification for each of the plurality of tile images, using the multiscale deep learning framework;
identifying, using the one or more processors, cells within each of the plurality of tile images using a trained cell segmentation model; and
identifying a predicted presence of one or more biomarkers associated with the digital image based at least in part on analyzing one or more tile images adjacent to the respective tile image and analyzing a larger tile image comprising a subset of the plurality of tile images, the subset containing the one or more tile images adjacent to the respective tile image,
wherein determining the tissue classification for each of the plurality of tile images comprises analyzing one or more tile images adjacent to the respective tile image and analyzing the larger tile image comprising the subset of the plurality of tile images and producing a 3D array of values that each represent the probability that a tissue class classification label matches a tissue classification in each tile image of the subset of the plurality of tile images.
|